REPLICATION
GENE:
The fragment of DNA which can be transcribed into functional RNAs like
r-RNA and t-RNA or translated into functional protein is termed as GENE.
GENOME:
The total genetic content of an unicellular
organism or single cell of an multicellular organism is known as genome.
CHROMOSOME STRUCTURE:
All types of nucleic acids interact with proteins. Chromosomal DNA forms stable nonspecific complexes with structural proteins that stabilize their tertiary structure; it also forms transient complexes with enzymes and regulatory proteins that modulate DNA and RNA metabolism. Chromosomal Structure of prokaryotes and eukaryotes are discussed below:
GENOME ORGANISATION IN
PROKARYOTES (CHROMOSOMAL STRUCTURE IN E.COLI):
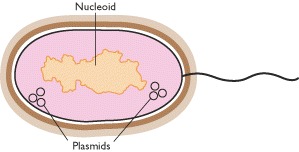
Prokaryotes including bacteria contain the genetic material in a fairly compact lump, occupying about one-third of the volume of the cell. Since the genetic material is not compartmentalized and lies naked in the cytoplasm, it is called nucleoid. The nucleoid is a folded structure, containing many supercoiled loops having many independent domains. There are about 100 domains per genome and each domain consists of about 40kb of DNA. The prokaryotic genome is a single replicon.
A single chromosome of E.Coli contains about 3x109 Daltons or about 4.5 x 106bp of DNA. If all of this DNA were in a duplex structure stretched end to end, it would be 1.5mm long, which is about 75 cell diameter. But inside the cell, the chromosome is coiled to the size of 2 micrometer. Electron micrographs of E.Coli chromosome suggest a folded circular structure containing 40-100 super coiled loops. This Structure is formed by the following steps i.e. initially circular DNA binds with the RNA-Protein core and forms loops. Secondly, the loops form super coiled structure. The interaction between the loops and RNA - Protein core is not understood. D.E. Pettijohn and his co-workers have provided evidence of such a core. They first showed that the individual supercoiled loops maintained their supercoiling independently of one another. Thus, if a single nick is introduced into one of the loops by limited DNase action. That loops adopts an expanded relaxed conformation, but supercoiling in the other loops is maintained. Limited RNase or protease treatment causes the partial breakdown of the looped structures without interfering with the supercoiling. These results have lead to the conclusion that each of the loops is a domain, the lateral motion of which is restricted by an RNA-Protein core complex.
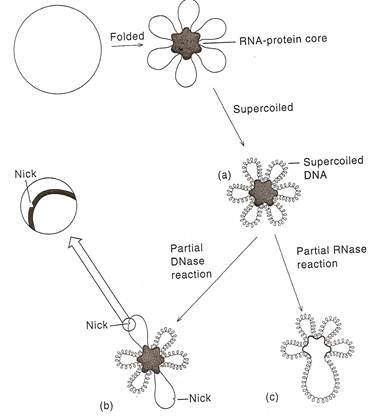
GENE ORGANIZATION IN PROKARYOTES:
The circular E.Coli chromosome contains enough base pairs to make about 3,000 average-sized genes. The relative positions of over half of these genes are known. The genes are present in a tightly organized form. Coding regions are interspersed with regulatory regions; there is no evidence for significant stretches of DNA with any function. Frequently, genes with a related function are clustered. These clustered genes are usually transcribed into single expression units (messenger RNA) containing the information for the synthesis of several functionally related proteins. Because of this character, in prokaryotes, genes are known as polycistronic in nature. The gene arrangement is explained with the concept of operons like Lac operon etc.
OPERONS:
One characteristic feature of prokaryotic genomes illustrated by E. coli is the presence of operons. An operon is a group of genes that are located adjacent to one another in the genome, with perhaps just one or two nucleotides between the end of one gene and the start of the next. All the genes in an operon are expressed as a single unit. This type of arrangement is common in prokaryotic genomes. A typical E. coli example is the lactose operon (catabolic operon), the first operon to be discovered (Jacob and Monod, 1961), which contains three genes involved in conversion of the disaccharide sugar lactose into its monosaccharide units - glucose and galactose. The monosaccharides are substrates for the energy-generating glycolytic pathway, so the function of the genes in the lactose operon is to convert lactose into a form that can be utilized by E. coli as an energy source. Lactose is not a common component of E. coli's natural environment, so most of the time the operon is not expressed and the enzymes for lactose utilization are not made by the bacterium. When lactose becomes available, it switches on the operon; all three genes are expressed together, resulting in coordinated synthesis of the lactose-utilizing enzymes.
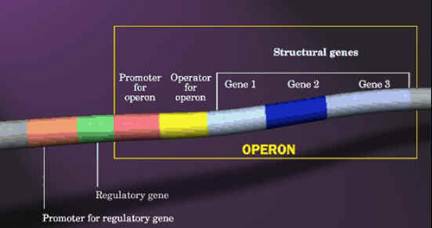
Similarly, there will be another operon namely Trp
operon which actively codes for enzymes which are involved in synthesis of
tryptophan (anabolic operon). By
considering the above two example one might think that operon will contain
genes for related metabolism but it is not true. In some cases like
the archaeon Methanococcus
jannaschii and the bacterium Aquifex
aeolicus have operons, but the genes in an
individual operon rarely have any biochemical relationship. For example, one of the operons in the A. aeolicus genome contains six linked genes, these genes
coding for two proteins involved in DNA recombination, an enzyme used in
protein synthesis, a protein required for motility, an enzyme involved in
nucleotide synthesis, and an enzyme for lipid synthesis. The main reason for
the presence of operons in prokaryotes provides more genes in a small region
due to the reduction in space for the presence of regulatory genes for each
gene separately.
EUKARYOTIC GENOME ORGANIZATION:
Eukaryotic DNA is contained in a relatively small number of chromosomes which varies according to the species. No direct correlation can be made between the amount of DNA in the nucleus and the number of chromosomes in which it is combined. Somatic cells of each species have two copies or homologous of each chromosome with the exception of the sex chromosome for which the female carries two 'X' chromosomes and the male an 'X' and a 'Y'. Germ cells contain only one copy of each chromosome and it is to these cells that the term haploid chromosome content or haploid DNA content refers. Aneuploid cells have an abnormal chromosome complement which is not necessarily on increase on the diploid condition for each chromosome type so that some may be present in greater numbers than others. The characteristic number and morphology of the chromosome in any particular cell type is known as the karyotype of that cell and is usually determined in metaphase when the chromosome are highly condensed and readily stained by basic dyes. Staining chromosome with basic dyes known as chromosome painting. In interpahase, the chromosomes are spread out in the nucleus and cannot be individually distinguished. Usually chromosome found to have “X” shape. Chromosome form of genome available only during Mitotic phase (division phase) whereas normally it present in Chromatin form in cells.

The centromere is the region of the chromosome to which spindle fibers attached. The centromere region usually appears to be constricted and the position of this constriction defines the ratio between the length of the two chromosome arms; this ratio is a useful characteristic centromere positions can be categorized as telocentric where centromere present at one end, acrocentric in which centromere is at off center, metacentric in which centromere present at centre and acentric in which chromosome lacks centromere.
Nucleoli are intranuclear organelles that contain rRNA, an important component of ribosomes. Different organisms are differently endowed with nucleoli, which range in number from one to many per chromosome set. The diploid cells of many species have two nucleoli. The nucleoli reside next to secondary constrictions of the chromosomes, called nucleolar organizers, which have highly specific positions in the chromosome set. Nucleolar organizers contain the genes that code for ribosomal RNA.
Chromomeres are beadlike, localized thickenings found along the chromosome during prophase of mitosis and meiosis. Homologous chromosomes tend to have homologous sets of chromomeres. Although they can be useful as markers, their molecular nature is not known.
When chromosomes are treated with chemicals that react with DNA, such as Feulgen stain, distinct regions with different staining characteristics are visually revealed. Densely staining regions are called heterochromatin; poorly staining regions are said to be euchromatin. The distinction refers to the degree of compactness or coiling, of the DNA in the chromosome. Heterochromatin can be either constitutive or Facultative. The constitutive type is a permanent feature of a specific chromosome location. The facultative type is sometimes but not always found at a particular chromosomal location. The pattern of heterochromatin and euchromatin along a chromosome are good cytogenetic markers. Chromatid is one of the two side by side replicas produced by chromosome division. Chromatin is the material of chromosomes, composed of DNA, chromosomal proteins and chromosomal RNA. Each eukaryotic chromosome contains a single, long, folded DNA molecule. Chromosomes responsible for sexual determination are generally named as allosomes (sex chromosomes) and others named as autosomes (somatic chromosomes). For example, in humans, XX or XY chromosomes considered as allosomes and other 22 pair chromosomes considered as autosomes.
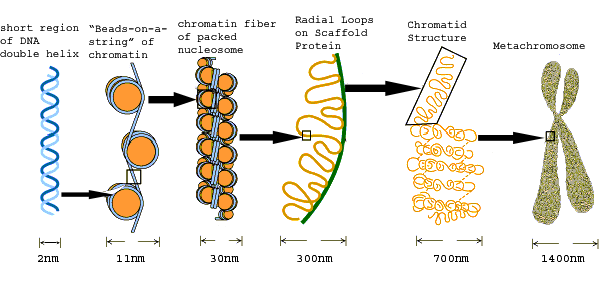
CHROMOSOME PACKING (CHROMATIN ORGANIZATION):
The Progressive levels of chromosome packing are as follows
i) DNA winds onto nucleosome spools
ii) The nucleosome chain coils into a solenoid
iii) The solenoid forms radial loops and the loops attach to a central scaffold
iv) The scaffold plus loops arrange themselves into a giant supercoil.
NUCLEOSOME [THE FIRST ORDER OF CHROMATIN FOLDING]:
The first level of chromatin folding was pointed out by Roger Kornberg in 1974. Nucleosome is the basic unit of eukaryotic chromosome. It is a ball of eight histone molecules wrapped about by two coils of DNA. Nucleosomes consists of the octomer Histones [H2A]2, [H2B]2, [H3]2 and [H4]2 in association with approximately 200bps of DNA. The fifth histone, H1, was postulated to be associated in some manner with the outside of the nucleosome. Nucleosome provide the extremely long thin chromosomal fiber with 20A'. The apparent decrease in length of the DNA in the minichromosome is due to the formation of nucleosomes. Infact, the DNA achieves about a seven fold condensation by coiling up into nucleosomes.
Each bead of nucleosome is actually a ball of histones with the DNA wound almost twice around the outside. Each ball contains exactly eight histone molecules, a pair each of H2A, H2B, H3 and H4. A single molecule of histone H1 binds to linker DNA outside the ball and can be removed by more stringent DNase treatment, which digests the linker DNA and yields a nucleosome core particle with about 150bps of DNA.
Nucleosome appears that the core histones are really not arranged in pairs. H3 and H4 form a tetramer at the center of the particle and these tetramers is flanked by H2A-H2B dimers at top left and lower right. Two properties of nucleosomes suggest that their function is fundamentally important. The first is the universality of nucleosomes in eukaryotes. The second is the extreme evolutionary conservation of most of the histones example: H4 from the cow differs only by two aminoacids from that of pea plant.
SOLENOID [SECOND ORDER OF CHROMATIN FOLDING]:
The next thickness, about 30nm (300A'), is due to further winding of the nucleosome to form a hollow coil called a solenoid.
Aaron Klug, who first described
the solenoid, traced its formation by making electron micrographs of chromatin
in solutions of increasing salt concentration. At very low salt
concentration, the chromatin appears as a string of nucleosomes. As the
salt concentration rises, coiling takes place, until the typical solenoid
structure appear. The DNA achieves another
RADIAL LOOPS [THIRD ORDER OF CHROMATIN FOLDING]:
Histone depleted metaphase chromosomes exhibit a central fibrous protein "scaffold" surrounded by an extensive hallow of DNA. The strands of DNA that can be followed are observed to form loops that enter and exit the scaffold at nearly the same point.
It is possible to digest 99% of the DNA from metaphase chromosome preparations leaving behind a morphologically intact central chromosome 'scaffold'. In a similar manner the material which remains when dehistonated interphase chromatin has most of its DNA removed by nuclease treatment is known as the nuclear 'matrix'. The relationship between the scaffold and matrix is unclear.
The scaffold proteins include two high molecular weight proteins of Mw of 1,70,000 and 1,35,000 but the matrix as well as containing an ill defined fibrous protein network is also associated with the nuclear pore lamina complex to which the chromosome are attached.
CHROMATID FORMATION [FOURTH ORDER OF CHROMATIN FOLDING]:
Most of these radial loops have lengths in the range 15 to 30 micrometer, so that when coiled as filaments they would be about 0.7micrometer long. Electron micrograph of a metaphase chromosome strongly suggests that the chromatid fibers of metaphase chromosomes are radially arranged. If the observed loops correspond to these radial fibers. They each contribute 0.3micrometer to the diameter of the chromosome. Taking into account the 0.4micron width of the scaffold, this model predicts the diameter of the chromatid of the metaphase chromosome to be 1 micron. Therefore, the total diameter of the chromosome will be around 1.4micron to 2 micron. A typical human chromosome, which contains ~ 140million bps, would therefore have 2000 of these 70kb radial loops.
EUKARYOTIC GENE ORGANIZATION:
According to the classical definition, a gene is a unique component of
the genome occupying a particular locus.
Each gene codes for a protein product and can be identified by mutations
that impede the protein function. Such
genes are structural genes. Another
class of genes is found in DNA that is in the form of multiple sequences. These genes code for the same protein or
closely related proteins. For the
purpose of identifying and characterizing structural genes, mRNA provides the
ideal intermediate and it can be subsequently used to translate into protein it codes for. It may
be noted that most structural genes lie in non-repetitive DNA. Structural genes may have three types of
organization:
a. They may consist of a DNA sequence
that code for a particular protein.
b. Besides structural genes, there
could be other sequences in the genome that code for related proteins –
repetitive and moderately repetitive.
c. Structural genes may be repeated,
i.e. there may be more than one copy of DNA sequence coding for a particular
protein.
The eukaryotic individual genes can be interrupted, there can be multiple
and identical copies of particular sequences and there may be large sequences
of nucleotides that do not encode for protein.
Another important feature of eukaryotes is the compartmentalization of
the nucleus and the cytoplasm that may create differences in the gene
expression.
The sequences of the DNA comprising the gene are thus divided into two
categories. The exons comprise of the
regions that are represented in the mRNA and used to produce the protein
product. The introns are missing from
the mRNA. The process of gene
expressions involves a new step, one that does not occur in bacteria. The DNA produces a copy of RNA with the exact
copy of gene sequence. This RNA is only
a precursor and in order to produce a functional mRNA, it undergoes splicing to
remove introns (non-coding sequences).
The eukaryotic gene structure is as follows:
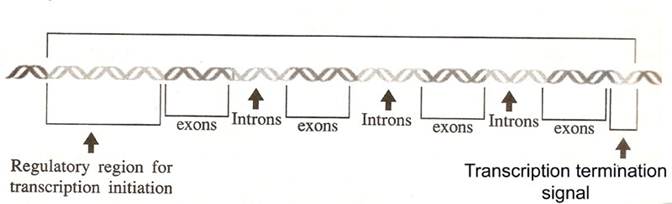
In eukaryotes, the relationship between gene and its RNA product is to be
understood and it is summarized as follows:
An interrupted gene consists of alternating series of exons and
introns. Exon sequences are present in
the RNA. Introns are intervening
sequences that are removed from the primary transcript and are absent from the
mature mRNA. A gene must start and end
with exons, corresponding to 5’ and 3’ ends of RNA.
An intervening sequence is an invariant feature. The intervening sequences vary enormously in
both number and size, but all classes of genes may be interrupted. The sequences of interrupted genes are same
as in case of mature RNA product. Thus genes split rather than be dispersed. Introns of nuclear genes generally have
termination codons in all reading frames and an interrupted gene retains the
same structure in all tissues. All
intervening sequences must be removed before they are translated. The length of the intron varies widely. They may be as short as 14 bp or as long as
46 bp in different genes. Introns are
non-repetitive and at the exon-intron junctions, consensus sequences are
located.
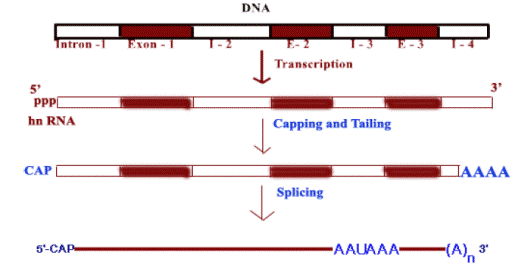
Eukaryotic
organelle genomes
The possibility that some genes might be located outside of the nucleus - extrachromosomal genes as they were initially called - was first raised in the 1950s as a means of explaining the unusual inheritance patterns of certain genes in the fungus Neurospora crassa, the yeast S. cerevisiae and the photosynthetic alga Chlamydomonas reinhardtii. Electron microscopy and biochemical studies at about the same time provided hints that DNA molecules might be present in mitochondria and chloroplasts. Eventually, in the early 1960s, these various lines of evidence were brought together and the existence of mitochondrial and chloroplast genomes, independent of and distinct from the nuclear genome, was accepted.
Origin of organelles
It is widely accepted that mitochondria and plastids evolved from bacteria that were engulfed by nucleated ancestral cells. As a relic of this evolutionary past, both types of organelles contain their own genomes, as well as their own biosynthetic machinery for making RNA and organelle proteins. Mitochondria and plastids are never made from scratch, but instead arise by the growth and division of an existing mitochondrion or plastid. On average, each organelle must double in mass in each cell generation and then be distributed into each daughter cell. Even nondividing cells must replenish organelles that are degraded as part of the continual process of organelle turnover, or produce additional organelles as the need arises. The process of organelle growth and proliferation is complicated because mitochondrial and plastid proteins are encoded in two places: the nuclear genome and the separate genomes harbored in the organelles themselves.
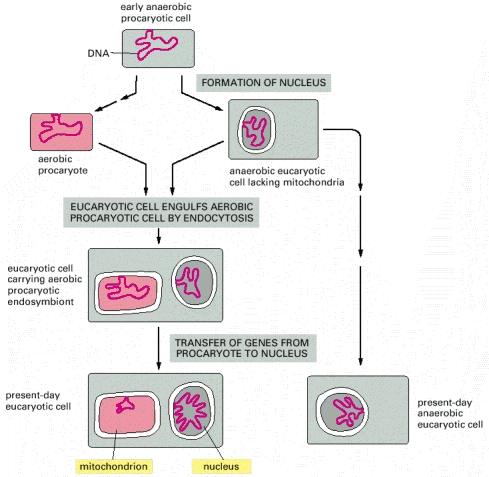
Microsporidia and Giardia are two present-day anaerobic single-celled eucaryotes (protozoans) without mitochondria. Because they have an rRNA sequence that suggests a great deal of evolutionary distance from all other known eucaryotes, it has been postulated that their ancestors were also anaerobic and resembled the eucaryote that first engulfed the precursors of mitochondria.
Physical features
of organelle genomes
Almost all eukaryotes have mitochondrial genomes, and all photosynthetic eukaryotes have chloroplast genomes. Initially, it was thought that virtually all organelle genomes were circular DNA molecules. Electron microscopy studies had shown both circular and linear DNA in some organelles, but it was assumed that the linear molecules were simply fragments of circular genomes that had become broken during preparation for electron microscopy. We still believe that most mitochondrial and chloroplast genomes are circular, but we now recognize that there is a great deal of variability in different organisms. In many eukaryotes the circular genomes coexist in the organelles with linear versions and, in the case of chloroplasts, with smaller circles that contain subcomponents of the genome as a whole. The latter pattern reaches its extreme in the marine algae called dinoflagellates, whose chloroplast genomes are split into many small circles, each containing just a single gene). We also now realize that the mitochondrial genomes of some microbial eukaryotes (e.g. Paramecium, Chlamydomonas and several yeasts) are always linear.
Copy numbers for organelle genomes are not particularly well understood. Each human mitochondrion contains about 10 identical molecules, which means that there are about 8000 per cell, but in S. cerevisiae the total number is probably smaller (less than 6500) even though there may be over 100 genomes per mitochondrion. Photosynthetic microorganisms such as Chlamydomonas have approximately 1000 chloroplast genomes per cell, about one-fifth the number present in a higher plant cell. One mystery, which dates back to the 1950s and has never been satisfactorily solved, is that when organelle genes are studied in genetic crosses the results suggest that there is just one copy of a mitochondrial or chloroplast genome per cell. This is clearly not the case but indicates that our understanding of the transmission of organelle genomes from parent to offspring is less than perfect.
Mitochondrial
genome
Mitochondrial genome sizes are variable and are unrelated to the complexity of the organism. Most multicellular animals have small mitochondrial genomes with a compact genetic organization, the genes being close together with little space between them. The human mitochondrial genome, at 16 569 bp, is typical of this type. Most lower eukaryotes such as S. cerevisiae, as well as flowering plants, have larger and less compact mitochondrial genomes, with a number of the genes containing introns.
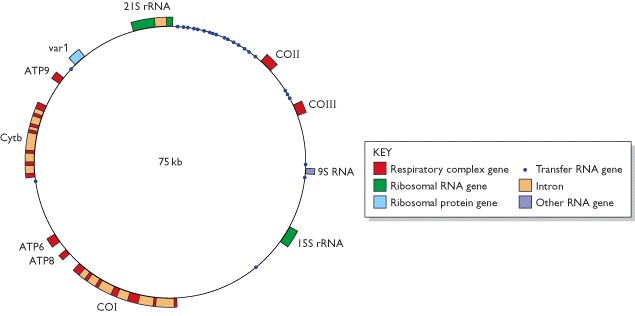
Because of their relatively small sizes, many mitochondrial genomes have been completely sequenced. In the yeast genome, the genes are more spaced out than in the human mitochondrial genome and some of the genes have introns. This type of organization is typical of many lower eukaryotes and plants. The yeast genome contains five additional open reading frames (not shown on this map) that have not yet been shown to code for functional gene products, and there are also several genes located within the introns of the discontinuous genes. Most of the latter code for maturase proteins involved in splicing the introns from the transcripts of these genes. Abbreviations: ATP6, ATP8, ATP9, genes for ATPase subunits 6, 8 and 9, respectively; COI, COII, COIII, genes for cytochrome c oxidase subunits I, II and III, respectively; Cytb, gene for apocytochrome b; var 1, gene for a ribosome-associated protein. Ribosomal RNA and transfer RNA are two types of non-coding RNA. The 9S RNA gene specifies the RNA component of the enzyme ribonuclease P.
Chloroplast
genomes
Chloroplast genomes have less variable sizes and most have a structure similar to that shown in figure for the rice chloroplast genome.
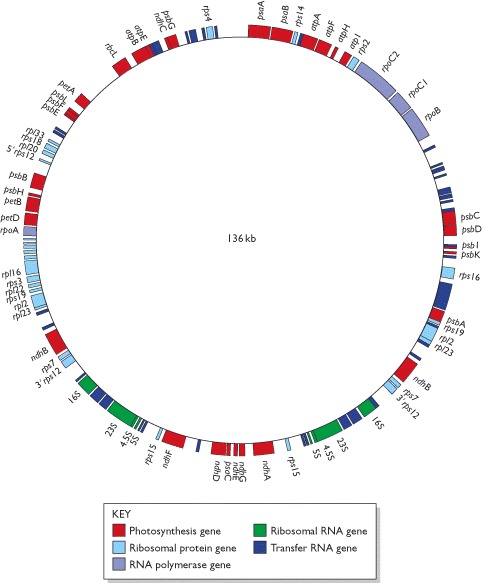
Only those genes with known functions are shown. A number of the genes contain introns which are not indicated on this map. These discontinuous genes include several of those for tRNAs, which is why the tRNA genes are of different lengths even though the tRNAs that they specify are all of similar size.
|
Species |
Type of organism |
Genome
size (kb) |
|
Mitochondrial genomes |
||
|
Plasmodium falciparum |
Protozoan (malaria parasite) |
6 |
|
Chlamydomonas reinhardtii |
Green alga |
16 |
|
Mus musculus |
Vertebrate (mouse) |
16 |
|
Homo sapiens |
Vertebrate (human) |
17 |
|
Metridium senile |
Invertebrate (sea anemone) |
17 |
|
Drosophila melanogaster |
Invertebrate (fruit fly) |
19 |
|
Chondrus crispus |
Red alga |
26 |
|
Aspergillus nidulans |
Ascomycete fungus |
33 |
|
Reclinomonas americana |
Protozoa |
69 |
|
Saccharomyces cerevisiae |
Yeast |
75 |
|
Suillus grisellus |
Basidiomycete fungus |
121 |
|
Brassica oleracea |
Flowering plant (cabbage) |
160 |
|
Arabidopsis thaliana |
Flowering plant (vetch) |
367 |
|
Zea mays |
Flowering plant (maize) |
570 |
|
Cucumis melo |
Flowering plant (melon) |
2500 |
|
Chloroplast genomes |
||
|
Pisum sativum |
Flowering plant (pea) |
120 |
|
Marchantia polymorpha |
Liverwort |
121 |
|
Oryza sativa |
Flowering plant (rice) |
136 |
|
Nicotiana tabacum |
Flowering plant (tobacco) |
156 |
|
Chlamydomonas reinhardtii |
Green alga |
195 |
Plasmids
A second complication concerns the precise status of plasmids with regard to the prokaryotic genome. A plasmid is a small piece of DNA, often, but not always circular, that coexists with the main chromosome in a bacterial cell. Some types of plasmid are able to integrate into the main genome, but others are thought to be permanently independent. Plasmids carry genes that are not usually present in the main chromosome, but in many cases these genes are non-essential to the bacterium, coding for characteristics such as antibiotic resistance, which the bacterium does not need if the environmental conditions are amenable. As well as this apparent dispensability, many plasmids are able to transfer from one cell to another, and the same plasmids are sometimes found in bacteria that belong to different species. These various features of plasmids suggest that they are independent entities and that in most cases the plasmid content of a prokaryotic cell should not be included in the definition of its genome.
|
Type of plasmid |
Gene functions |
Examples |
|
Resistance |
Antibiotic resistance |
Rbk of Escherichia coli and other bacteria |
|
Fertility |
Conjugation and DNA transfer between bacteria |
F of E. coli |
|
Killer |
Synthesis of toxins that kill other bacteria |
Col of E. coli, for colicin production |
|
Degradative |
Enzymes for metabolism of unusual molecules |
TOL of Pseudomonas putida, for toluene metabilism |
|
Virulence |
Pathogenicity |
Ti of Agrobacterium tumefaciens, conferring the ability to cause crown gall disease on dicotyledonous plants |
Genes carried by plasmids are useful, coding for properties such as antibiotic resistance or the ability to utilize complex compounds such as toluene as a carbon source, but plasmids appear to be dispensable - a prokaryote can exist quite effectively without them.
Plasmids are circular molecules of DNA that lead an independent existence in the bacterial cell. Plasmids almost always carry one or more genes, and often these genes are responsible for a useful characteristic displayed by the host bacterium. For example, the ability to survive in normally toxic concentrations of antibiotics such as chloramphenicol or ampicillin is often due to the presence in the bacterium of a plasmid carrying antibiotic resistance genes. In the laboratory, antibiotic resistance is often used as a selectable marker to ensure that bacteria in a culture contain a particular plasmid. Most plasmids possess at least one DNA sequence that can act as an origin of replication, so they are able to multiply within the cell independently of the main bacterial chromosome. The smaller plasmids make use of the host cell’s own DNA replicative enzymes in order to make copies of themselves, whereas some of the larger ones carry genes that code for special enzymes that are specific for plasmid replication. A few types of plasmid are also able to replicate by inserting themselves into the bacterial chromosome. These integrative plasmids or episomes may be stably
maintained in this form through numerous cell divisions, but always at some stage exist as independent elements.
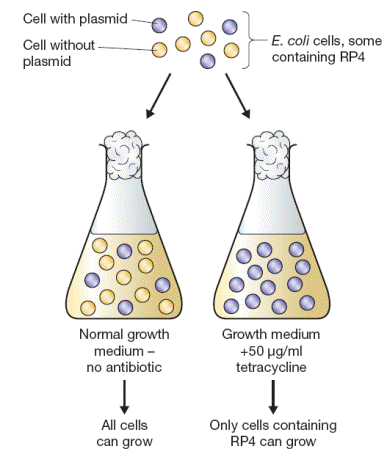

The copy number refers to the number of molecules of an individual plasmid that are normally found in a single bacterial cell. The factors that control copy number are not well understood. Some plasmids, especially the larger ones, are stringent and have a low copy number of perhaps just one or two per cell; others, called relaxed plasmids, are present in multiple copies of 50 or more per cell. Generally speaking, a useful cloning vector needs to be present in the cell in multiple copies so that large quantities of the recombinant DNA molecule can be obtained.
Conjugative plasmids are characterized by the ability to promote sexual conjugation between bacterial cells Several different kinds of plasmid may be found in a single cell, including more than one different conjugative plasmid at any one time. In fact, cells of E. coli have been known to contain up to seven different plasmids at once. To be able to coexist in the same cell, different plasmids must be compatible. If two plasmids are incompatible then one or the other will be rapidly lost from the cell. Different types of plasmid can therefore be assigned to different incompatibility groups on the basis of whether or not they can coexist, and plasmids from a single incompatibility group are often related to each other in various ways. The basis of incompatibility is not well understood, but events during plasmid replication are thought to underlie the phenomenon.
Although plasmids are widespread in bacteria they are by no means as common in other organisms. The best characterized eukaryotic plasmid is the 2 mm circle that occurs in many strains of the yeast Saccharomyces cerevisiae. The discovery of the 2 mm plasmid was very fortuitous as it allowed the construction of cloning vectors for this very important industrial organism. However, the search for plasmids in other eukaryotes (such as filamentous fungi, plants and animals) has proved disappointing, and it is suspected that many higher organisms simply do not harbor plasmids within their cells.
MODES OF REPLICATION
There are three modes are proposed for replication by Crick namely Conservative mode, Semiconservative mode and Dispersive mode. Of the three, Semiconservative mode of replication in prokaryotes proved by Meselson-Stahl experiment.
WATSON AND CRICK MODEL OR HYPOTHESIS ON REPLICATION:
The Watson and Crick model for DNA replication assumes that as new strands of DNA are made, they follows the usual base pairing rules of A with T and G with C. This is essential because the DNA replicating machinery must be capable of discerning a good pair from a bad one, and the Watson - Crick base pairs give the best fit. Three hypotheses are available for DNA replication. They are
i) Conservative replication
ii) Semiconservative replication
iii) Dispersive replication
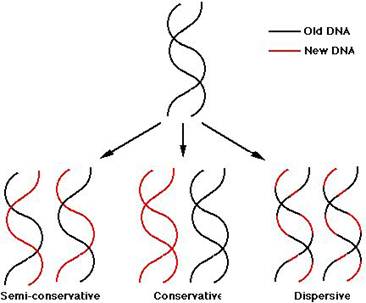
Conservative replication: It yields two daughter duplexes, one of which has two old strands and one of which has two new strands.
Semiconservative replication: It gives two daughter duplex DNAs, each of which contains one old strand and one new strand.
Dispersive replication: It yields two daughter duplexes, each of which contains strands that are mixture of old and new DNA.
EVIDENCE FOR SEMI CONSERVATIVE REPLICATION:
In 1958, Matthew Meselson and Franklin Stahl worked out clever procedure to distinguish semi conservative DNA replication from conservative or dispersive replication, using a nonradioactive heavy isotope of nitrogen. Ordinary nitrogen, the most abundant isotope, has an atomic weight of 14, so it is called 14N. A relatively rare isotope 15N has an atomic weight of 15. Meselson and Stahl found that if bacteria grow in a medium enriched in 15N, they incorporate the heavy isotope into their DNA, which becomes denser than normal. This labeled DNA CLEARLY separates from ordinary DNA in gradient of Cesium Chloride (CsCl) spun in an ultracentrifuge. CsCl is used because it is a very dense salt and therefore makes dense enough solution that DNA will float somewhere in the middle rather than sinking to the bottom.
AIM:
The crux or aim of the experiment was to grow 15N labeled bacteria in 14N-medium for one or more generations and then to look at the density of the DNA products.
RULING OUT CONSERVATIVE MODEL:
Consider the different outcomes we would expect after one round of replication according to the three different mechanisms. If replication is conservative, the two heavy parental strands will stay together and another, newly made DNA duplex will appear. Since this second duplex will be made in the presence of light isotope, both its strands will be light. The heavy / heavy parental duplex and light / light progeny duplex will separate readily in the CsCl gradient. On the other hand, if replication is semi conservative or dispersive, the two heavy parental stands will separate and each will be supplied with a new light partner and heavy and light strand dispersed randomly between parental and progeny strands respectively. These heavy / heavy hybrid duplexes will have a density halfway between the heavy / heavy parental DNA and light / light ordinary DNA. Meselson & Stahl results confirmed the latter prediction, so the conservative mechanism ruled out.
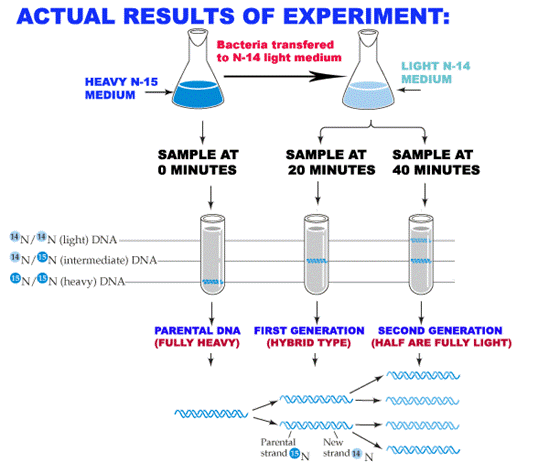
RULING OUT RANDOM DISPERSIVE MODEL:
The results of one more round of DNA replication destroyed the random dispersive hypothesis. The random dispersive model predicts that the DNA after the second generation will have a single density, corresponding to molecules that are 25% heavy and 75% light. This should give one band of DNA halfway between light / light band and the heavy / light band. The semi conservative model predicts equal amounts of two different DNAs. Again, the latter prediction matched the experimental results, confirming the semi conservative model.
RULING
OUT NON-RANDOM DISPERSIVE MODEL:
Nonrandom dispersive distribution would yield the same gradient centrifugation result as would semi conservative replication after two rounds of replication. To eliminate the possibility of such dispersive replication, Meselson and Stahl took DNA after one round of replication and heated it to separate its stands. Then they subjected the separated strands to gradient centrifugation. The semi conservative model predicts that half the strands should be heavy and half should be light. These should form separate bands upon centrifugation coincident with standards prepared from pure heavy and light DNAs. On the other hand, any dispersive model predicts that both stands should be mixed and therefore should not form bands coincident with pure heavy and light DNA strands.
In Meselson and Stahl's experiment, the two strands were readily separable, and behaved completely light and completely heavy DNA strands, just as the semi conservative model demands. This allows us to rule out the nonrandom dispersive hypothesis.
CONCLUSION:
DNA replicates in a semi conservative manner. When the parental strands separate, each serves as the template for making a new, complementary strand.
AUTORADIOGRAPHIC DEMONSTRATION OF DNA
REPLICATION (John
The Semiconservative method of replication
was verified photographically in 1963 by J.Cairns who
used the technique of autoradiography. This technique makes use of the
fact that radioactive atoms expose photographic film. The visible silver grains
on the film can then be counted to provide an estimate of the quantity of
radioactive material present.
The first, known at the time, is that the E.Coli DNA is a circle. The Second point is that DNA is replicated while maintaining the integrity of the circle i.e., the circle does not appear to be broken in the process of DNA replication; an intermediate theta structure is formed which is due to the formation of replication eye. Third, replication of the DNA seems to be occurring at one or two moving Y-junctions in the circle Replication forks, which further supports the Semiconservative replication.
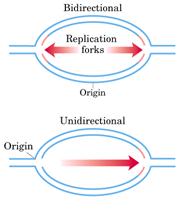
REPLICATION IS BI-DIRECTIONAL:
In some cases, radioactivity had not been
applied to the cell until DNA replication had already begun. In these cases,
the radioactive label appeared after the theta structure had already begun
forming. After some time, the process was stopped and the
autoradiographs,
Result:
As the grain track pattern present on both
forks with unlabeled DNA in the middle, it was concluded that the direction of
DNA synthesis is bi-directional. The
unidirectional replication was ruled out because had it been the case, the
grain tracks should have been on only one side of the point of origin.
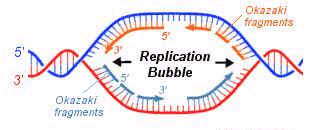
REIJI
OKAZAKI EXPERIMENT (Semidiscontinuous Replication):
Autoradiograph images of replicating DNA of E.Coli suggest that DNA's two antiparallel strands are simultaneously replicated as the replicating fork advances in diametrically opposite directions.
All known DNA polymerases catalyze the chain growth only in 5'--> 3' direction. Therefore, synthesizing daughter DNA on 3’--> 5' template is done by polymerase as the 3' group is free for the chain growth in 5'-->3' direction. But how can polymerase catalyze replication on template of 3'-->5'. In this case only 5' end would be available, polymerase not use it to start the chain growth. Reiji Okazaki’s Pulse-Chase Experiment answered this baffling question.
1) Since atleast half of the newly synthesized DNA appears first as short pieces, it is necessary to fix the short pieces of DNA by labeling them with radioactive precursors. These short bursts of labeling with radioactive substances are called pulses. The stopping period is called chase.
2) If we eliminate the enzyme (DNA ligase) that is responsible for Stiching together the short pieces of DNA, we ought to too be able to see these pieces even with long pulses of DNA precursor.
For his model system,
To test first prediction,
REPLICATION EYE:
It is formed during replication in both eukaryotic and prokaryotic DNA. It is the place where replication occurs actively. It is otherwise known as replication bubble. Formation of the replication eye provides the theta like structure to the circular DNA during replication in prokaryotes. Each replication bubble found to have two replication forks, each at the corner of an eye.
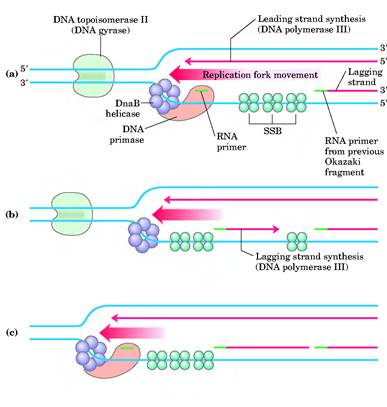
REPLICATION FORK:
The corner region of the replication bubble or replication eye is referred as replication fork. Generally it is point from which the replication elongated or continued. Bi-directional nature of replication was identified by studying the appearance of nucleotides at the replication forks in connection with the change in the radioactive material intensity. Each replication bubble found to have two replication forks, each at the corner of an eye.
SEMIDISCONTINUOUS
REPLICATION:
During Replication, half of the DNA
strand synthesized in fragments where as remaining half synthesized
continuously. This is referred as
semidiscontinuous replication. This happens
only because of the nature of the DNA polymerases i.e.,
they can synthesize DNA by extending 3’end and they are unable to extend DNA
from 5’end. This nature of replication
is proved by
Fragments synthesized during lagging
strand formation of replication was identified and proved by Rejis
Okazaki. Hence, by his name these
fragments are called as
RNA
PRIMERS:
RNA primers are single strand oligoribonucleotides with 40 to 60 base pairs. For replication RNA primers are required. This is because DNA polymerases require RNA primer for their action to start in invivo condition. In replication RNA primer is synthesized by two different enzymes namely RNA polymerase and Primosome complex. RNA polymerase synthesize RNA primer for synthesize of leading strand where as Primosome synthesize RNA primer for lagging strand synthesis. In addition to this, RNA polymerase synthesizes only one primer whereas Primosome synthesizes many primers.
ENZYMES OF REPLICATION:
DNA DIRECTED DNA POLYMERASES:
This enzyme synthesizes DNA by utilizing DNA template.
PROKARYOTIC DNA DIRECTED DNA POLYMERASES:
In prokaryotes, there are three kinds of DNA polymerases. They are
I. DNA directed DNA polymerase - I
II. DNA directed DNA polymerase - II
III. DNA directed DNA polymerase - III
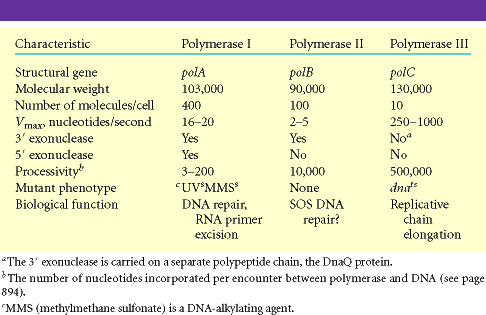
I. DNA directed DNA polymerase - I (Kornberg enzyme):
Its Molecular weight is 109kd and the concentration is around 400 molecules per cell. When treated with proteases like Trypsin, it gets cleaved into two fragments possessing different function. The large fragment called klenow fragment posses both polymerase and 3'-->5' exonuclease activity. The small fragment possesses only 5'-->3' exonuclease activity. Overall, three activities are possessed by DNA Pol-I.
i) DNA polymerase activity
ii) 3'--> 5' exonuclease activity
iii) 5'-->3' exonuclease activity
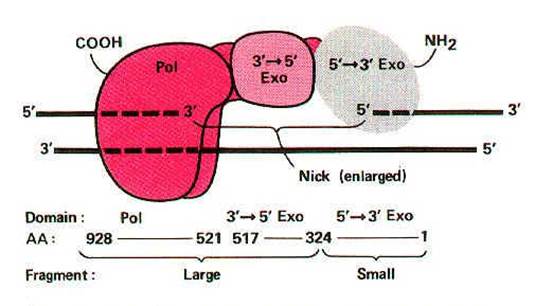
i) DNA polymerase activity:
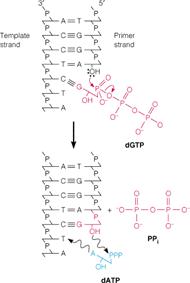
It catalyzes synthesis of complementary DNA strand from the appropriate deoxy nucleotides using single or double stranded DNA as template and DNA or RNA as primer. DNA polymerases catalyze polymerization reaction, which involves step-by-step addition of deoxy nucleotides unit to a DNA or RNA primer chain. It is represented by equation
n(dNTPs)--->(dNMP)n
The chain elongation reaction catalyzed by DNA polymerase occurs by means of a nucleophilic attack of the 3'-OH terminus of the primer on the alpha phosphorous atom of the incoming deoxyribonucleotide and releasing pyrophosphate molecule. The subsequent hydrolysis of "ppi" drives the reaction in the forward direction. It is represented in the following figure:
CONDITIONS FOR DNA POLYMERASE ACTION:
a) All four dNTPs (dATP, dGTP, dTTP, dCTP) along with Mg2+ ions.
b) A primer chain with a free 3'-OH group. Invitro conditions require DNA primer strand while invivo condition require RNA primer strand. Generally DNA Pol – I utilizes DNA primer whereas DNA Pol – III utilizes RNA Primer.
c) A DNA template, which can be single stranded or double stranded DNA. Double stranded DNA is an effective template only if its sugar phosphate backbone is broken at one or more sites.
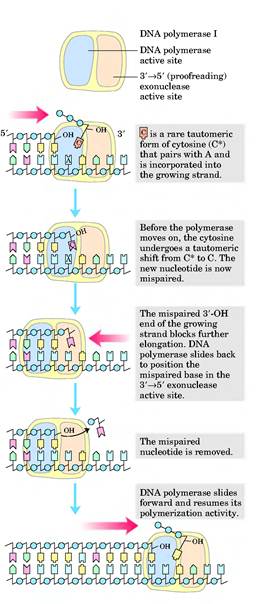
ii) 3'-->5' exonuclease activity:
This is the proofreading or editing activity of DNA polymerase, which gets activated when a wrong base pair is incorporated into the chain. DNA pol-I removes deoxy nucleotide at 3'-end of the progeny DNA. Because of this, the activity is referred as 3'-->5' exonuclease activity. Due to this activity, it repairs DNA during replication.
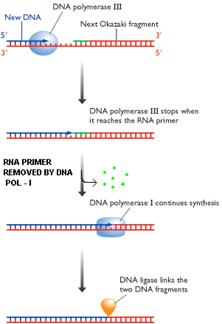
iii) 5'-->3' exonuclease activity:
With the help of this activity, enzyme removes nucleotides from 5'-end. Two functions can be carried out by this activity. They are removal of RNA primer and DNA repair.
a)
Removal of Primer:
b) DNA repair:
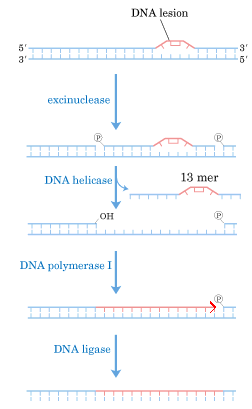
II) DNA directed DNA Polymerase-II: structure and function are not completely elucidated. It has polymerase activity along with proof reading function, but it does not have 5'-->3' exonuclease activity.
III) DNA directed DNA Polymerase-III:
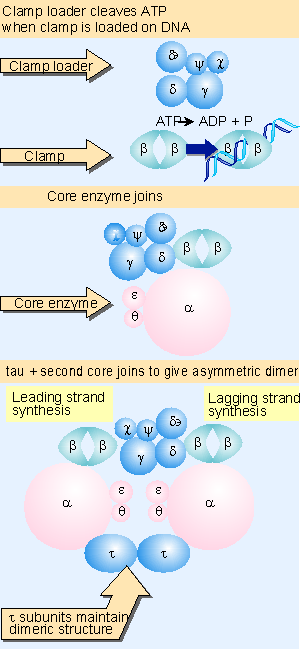
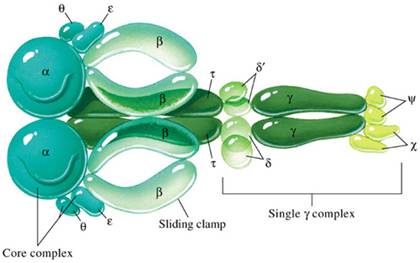
Holoenzyme of DNA pol-III consists of ten subunits. They are aeqbgdd'cjt . Core enzyme of DNA poly-III contain three subunit aeq. Of three subunits alpha subunit posses polymerizing activity and epsilon posses proof reading activity. These are the two function of the enzyme. It does not have 5'-->3' exonuclease activity.
|
S.No |
SUBUNITS |
FUNCTION |
|
1. |
a |
Polymerization |
|
2. |
e |
Repair and Proof reading function |
|
3. |
q |
Assembly of DNA pol-III |
|
4. |
b |
Processivity - It holds the template |
|
5. |
gdd' |
Lagging strand synthesis - They aid the formation of loop. |
|
6. |
cj |
Normal function of DNA pol-III |
|
7. |
t |
Dimerization |
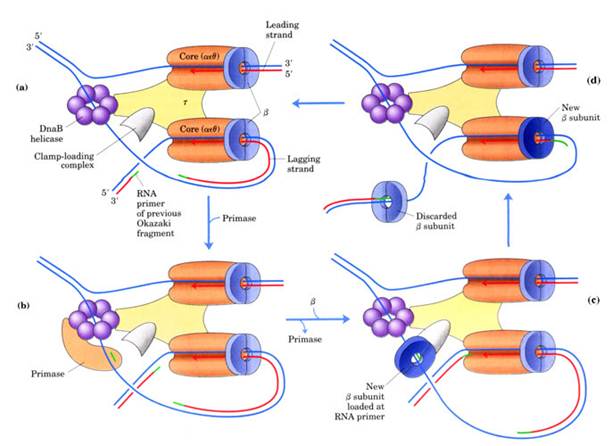
gdd'c and j subunits called as a gamma complex, which mediates transfer of the beta subunit to the duplex DNA primer strand. DNA pol-III possesses high processivity than other DNA polymerases. This is because of beta subunit. It form a donut shaped dimers around duplex DNA and then associate with and hold the catalytic core aeq polymerase at the primer template terminus.
Once tightly associated with DNA, the beta subunit dimers functions like a "clamp" which can move freely along the DNA, like a ring on a string carrying the associated core polymerase with it. In this way, the active sites remain near the growing fork and the processivity of the core polymerase maximized.
Processivity: Number of nucleotides added by the enzyme before it gets released from the template strand after its attachment.
EUKARYOTIC DNA DIRECTED DNA POLYMERASE:
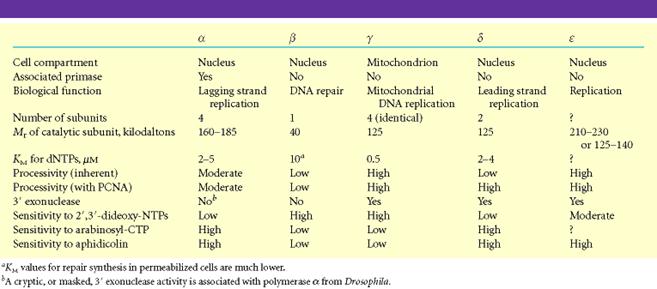
Following are the six important eukaryotic DNA polymerases. They are as follows:
i) DNA polymerase a
ii) DNA polymerase b
iii) DNA polymerase g
iv) DNA polymerase d
v) DNA polymerase e
vi) DNA polymerase s
i) DNA polymerase a
It is composed of four subunits of which one has the primase activity. The largest subunit of it has polymerase activity. It is supposed to exist in several forms like a1,a2, and a3. DNA polymerase a in association with DNA polymerase d , is involved in the replication of nuclear chromosomal DNA. The replication is also aided by another protein factors called Accessory Proteins and AP4A. These protein factors appear to have regulatory function. DNA pol a is supposed to carryout lagging strand synthesis.
ii) DNA polymerase b:
It is involved in the repair of DNA.
iii) DNA polymerase g:
This polymerase is involved in the replication of mitochondria DNA.
iv) DNA polymerase d:
This polymerase has polymerase function and 3'-->5’ exonuclease activity. It is involved in leading strand synthesis.
v) DNA polymerase e:
It is supposed to replace DNA pol delta in some situations such as DNA repair.
vi) DNA polymerase s:
This polymerase seems to be expressed only in the bone marrow cells. It is the only eukaryotic polymerase that has a deoxy ribonuclease activity.
DNA LIGASE:
The DNA ligases are responsible for connecting DNA segments during replication, repair and recombination. They are class of enzymes that catalyze the formation of alpha-phosphodiester bond between two DNA chains. This enzyme requires the free OH group at the 3' end of other DNA strand and phosphate group at 5' end of the other. The formation of a phosphodiester bond between these groups is an endergonic reaction. Hence energy source required for ligation. In E.Coli and other bacteria NAD+ supplies the energy whereas in animals i.e. eukaryotes ATP play the role.
Mechanism:
a) The adenosyl group of the ATP or NAD+ is transferred to the enzyme ligase to form covalent enzyme-AMP complex in which the AMP is linked to the epsilon amino group of lysine residue of the enzyme through posphamide bond.
b) The enzyme then transfers the adenyl group to the 5' phosphoryl terminus of the nick to form a pyrophosphate grouping. In effect, AMP is attached to the 5'-phosphoryl terminus.
c) The final step is a nucleophilic attack of the 3'-OH group on the 5' activated phosphorus atom. A phosphodiester bond is formed and AMP is released. This reaction is driven by the hydrolysis of the pyrophosphate group. Thus two high-energy phosphate bonds are spent in forming a phosphodiester bridge in the DNA backbone.
Role of DNA ligases in Replication:
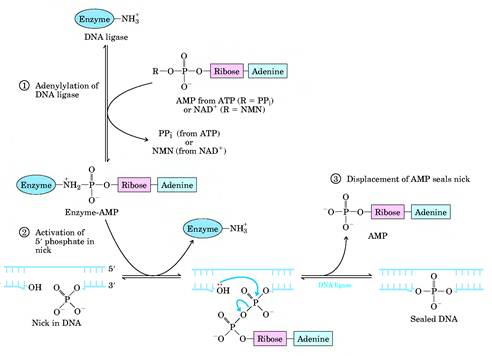
This enzyme seals the nick and joints
the
DNA HELICASES:
They are proteins, which are involved in the unwinding of DNA molecule. There are four kinds of helicases namely Dna B, Rep Proteins, Helicases - II and Dna A.
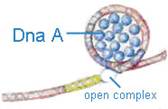
Dna-A protein (Mw 48,000) - It binds to 4 of 9mer sequence and unwinds a 3 of 13mer sequence at Ori C site and forms an open complex during initiation of replication. It is the first protein which binds to DNA to initiate DNA replication.
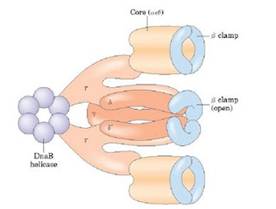
Dna-B proteins (Mw- 3,00,000)- It is a primosome constituent and consists of six subunits. It unwinds DNA during replication. It is responsible for the extension of open complex during replication.
Rep Proteins (Mw 65,000) - It is a helicase consisting of one subunit. It binds to the 5'3'template and moves in 3'5' direction. It actively participates in leading strand synthesis in replication.
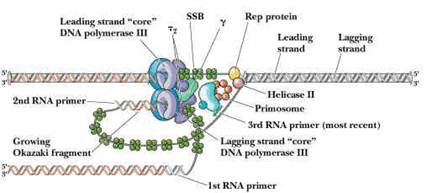
DNA Helicase - II (Mw 75,000) - It is helicase consisting of only one subunit. It binds with 3'5' template stand and moves along in 5'3' direction. It is involved in lagging strand synthesis in replication.
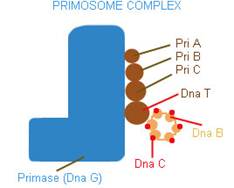
PRIMOSOME:
It is a complex containing Pri A, Pri B, Pri C, Dna T, Dna B, Dna C and Primase (Dna G) proteins. Primososme complex without primase referred as preprimosome complex. Primosome complex plays a vital role in lagging strand synthesis by synthesizing the primer at frequent intervals. Of all the proteins actual primer synthesizing ability relies with primase molecule. But each protein of the complex have their own role i.e. each of them necessary for the function of primosome complex in replication for example. Pri A , Pri B, Pri C necessary for the binding of primosome complex to DNA. Dna B responsible for helicase activity. Dna C aids the binding of Dna B. Dna T helps the binding of DnaB and Dna C complex to Pri A, Pri B and Pri C complex.
SUPERCOILING
OF DNA:
When compare to the size of the cells (both in eukaryotes and prokaryotes) the dimensions i.e. the contour length of DNA, is colossally large. The DNA is packed into the cells not by mere folding and stuffing it in a random way but rather in a highly organized fashion as it has to provide an access in a very systematic manner to a vast array of enzymes and other factors that control its replication, transcription and gene regulation. Therefore DNA is accommodated into other small space by an organized bending or twisting of the molecule into a structure called supercoiled structure.
DNA is coiled around an axis in the forms of a double helix and the bending or twisting of that access upon itself is refers to as DNA supercoiling. Therefore, supercoiling represents twisting or bending of the DNA double helix upon itself. When there is no bending or twisting of DNA axis upon itself, the DNA is said to be in a relaxed state. Supercoiling may occur in two different structural forms, they are intertwined form or toroidal form. In intertwined form, DNA strand twisted upon itself without any support. In toroidal form, it twisted upon a component ex: proteins. Supercoiling of DNA is of two types namely Positive supercoiling and Negative supercoiling.

POSITIVIE SUPERCOILING:
If a circular DNA molecule in a relaxed conformation is broken across both strands and one or more additional right handed helical turns are inserted before rejoining the ends. The molecule would twist on itself to form positive super coil.
NEGATIVE SUPERCOILING:
If a circular DNA molecule in a relaxed conformation given a double strand break and unwound by giving left handed helical turn and the ends rejoin, the molecule will twist itself to form negative supercoil.
These two forms of DNA, which will only differ in their topological structures, are called topoisomers.
Terms:
Twist: Number of turns by one-polynucleotide chains around another on double helical axis is referred as twist (t).
Writh: Number of turns by double helical DNA upon itself or around central axis referred as writh (w).
Linking Number: In circular DNA, Linking number (LK) specifies total number of turns that are present in a closed circular DNA. Otherwise, it is a sum of twist and writh.
L= t+ w
Linking Number is a constant property and it can be changed only by phosphodiester bond breaking. If the linking number is reduced by the process of unwinding in the relaxed circular DNA, it induces negative supercoiling. If X is the number of turns in the relaxed circular DNA (Lk0=x) and if one turn is removed by unwinding (LK=X-1), it induces negative supercoiling.
Therefore, the change in linking number DLK= LK - LK0
=X - 1 - X= -1
DLK = -1
On the other hand if one turn is added to the relaxed circular DNA, if result in positive supercoiling, therefore the change in linking number DLK is +1.
TOPOISOMERASES:
The normal biological function of DNA occurs only if it is in proper topological state, i.e. to stay in having proper supercoiling or super helical tension. Topoismerases are a group of enzymes which controls supercoiling of DNA thereby maintaining it in the proper topological state or superhelical tension. There are two classes of topoisomerases. They are type – I topoisomerase and type – II topoisomerase.
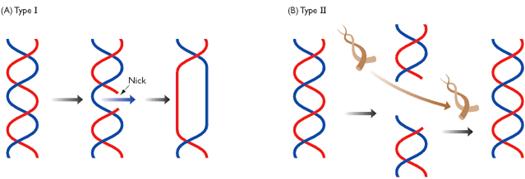
TYPE - I TOPOISOMERASE:
Type-I topoisomerase (Nicking-Closing enzymes) are monomeric 100kd proteins that are widespread in both prokaryotes and eukaryotes. They can remove negative supercoils without leaving nicks in the DNA molecule.
Mechanism:
After the enzyme binds to a DNA molecule and cuts on strand, the free 5' phosphate on the DNA is covalently attached to a tyrosine residue in the enzyme in the case of prokaryotes (the free 3'-phosphate on the DNA is covalently attached in the case of eukaryotes). The DNA strand that has not been cleaved is then passed through the single stranded break. The cleaved strand is then resealed. By this mechanism, the enzyme removes one negative supercoil at a time there by increasing the LK by one in the case of prokaryotes. By forming free 3' or free 5' end DNA- protein covalent intermediates, the free energy of the cleaved phosphodiester bond is preserved so that no energy input is required to reseal the nick.
The type -I topoisomerase from E.Coli acts on negatively supercoiled molecules but not on positively supercoiled molecules. In contrast, type-I topoisomerase from eukaryotic cells can remove both positive and negative supercoils. Type-I topoisomerase reversibly catenates (interlinks) single stranded circles. Topoisomerases-I and Topoisomerases-II are type-I topoisomerases.
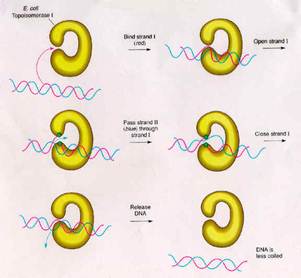
TYPE-II TOPOISOMERASES:
Prokaryotic type-II topoisomerases, which are also known as DNA gyrases, are 375kd proteins that consists of two pairs of subunits designated A and B. DNA gyrase converts a right-handed toroidal supercoil to a left-handed toroidal supercoil. This mechanism is called as sign inversion mechanism. DNA gyrase requires ATP for its function. Hydrolysis of ATP is not required to induce supercoils but is required for the enzyme to turnover and introduces additional supercoils.
The type-II topoisomerase enzymes from mammalian cells cannot increase the superhelicity (super helical density) at the expense of ATP, presumably no such activity is required in eukaryotes since binding of histones increases the potential superhelicity. DNA gyrase has the ability to cut a double stranded DNA molecule, pass another portion of the duplex DNA through the cut, and reseal the cut. It changes the linking number of the DNA by 2.
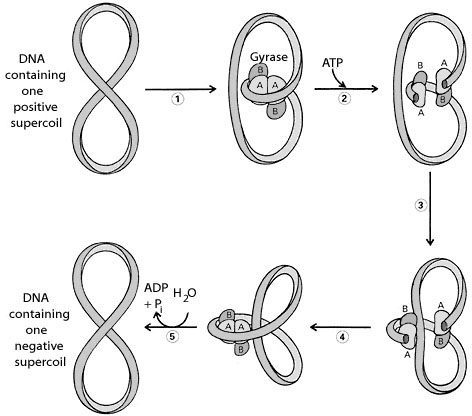
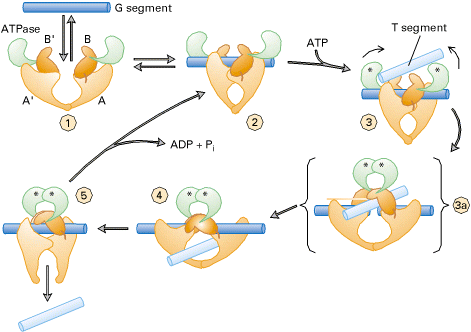
|
S.No |
Properties |
Prokaryotes |
Eukaryotes |
||
|
Type-I |
Type-II |
Type-I |
Type-II |
||
|
1. |
Molecular Weight |
100,000 |
400,000 |
1,00,000 |
3,09,000 |
|
2. |
Subunits |
Monomer |
Tetramer |
- |
Dimers |
|
3. |
Gene |
Topo - A |
gyr - A gyr - B |
- |
- |
|
4. |
Covalent Intermediate DNA |
At 5'-P |
Subunit A at 5'-P |
At 3'-P |
At 5'- P |
|
5. |
Relaxation |
Mg2+ dependent ATP independent |
ATP dependent |
Mg2+ independent ATP independent |
ATP dependent |
|
6. |
Negative supercoiling |
- |
ATP dependent |
- |
- |
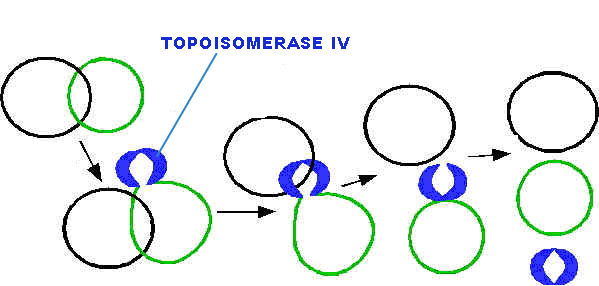
All type-II topoisomerase catalyze catenation and decatenation that is, the linking and unlinking of two different DNA duplexes. Topoisomerase-IV is also a type of type-II topoisomerases. It actively involved in decatenation i.e. termination of DNA replication.
SINGLE
Single
strand binding protein named because its nature to bind to single strand of DNA
which will be formed during open complex formation during initiation of
replication. It contains four subunits. The
major function of SSB is to prevent recoiling of DNA strands after it’s
unwinding by helicases. Thus, SSB plays
vital role in replication.
REPLICATION IN PROKARYOTES
Replication is an enzymatic process in
which synthesis of a daughter or progeny duplex DNA molecule, identical to the
parental duplex DNA occurs. Rate of
replication in E.Coli
(prokaryotic cell) is 1500 nucleotides per second. To complete replication of whole E.Coli genome it takes 40
minutes. Rate of replication in eukaryotes is about 10 - 100 nucleotides
per second. To complete replication of simple eukaryotic genome 6 - 8
hours required. In prokaryotic circular DNA only one replication
fork is present but in eukaryotic DNA several replication forks are
present. Space between two-replication forks in eukaryotes is about
20kbps apart.
THE REPLICATION OF CIRCULAR DNA IN E.COLI (Prokaryotic duplex DNA replication) (q REPLICATION):
The synthesis or replication of DNA molecule can be divided into three stages
I) Initiation (Formation of Replisome)
II) Elongation (Initiation of synthesis and elongation)
III) Termination
I) Initiation
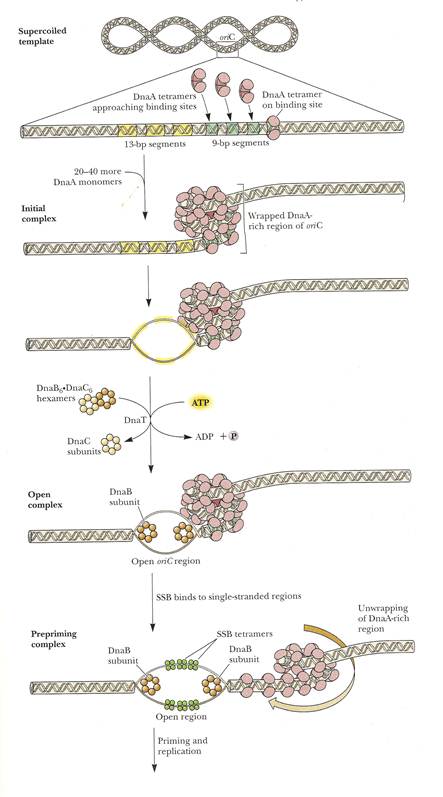
The replication begins at a specific initiation point called OriC point or replicon. (Replicon: It is a unit of the genome in which DNA is replicated; it contains an origin for initiation of replication) It is the point of DNA open up and form open complex leading to the formation of prepriming complex to initiate replication process.
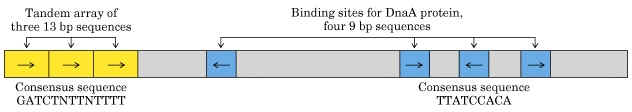
The OriC site is situated at 74" minute near the ilv gene. The OriC site consists of 245 basepairs, of which three of 13 basepair sequence are highly conserved in many bacteria and forms the consensus sequences (GATCTNTTNTTTT). Close to OriC site, there are four of 9 basepair sequences each (TTATCCACA).
The sequence of reactions in the initiation process is as follows:
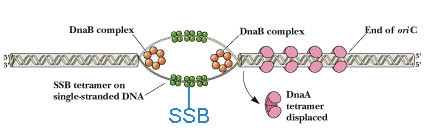
a) Dna A protein recognizes and binds up to four 9bp repeats in OriC to form a complex of negatively supercoiled OriC DNA wrapped around a central core of Dna A protein monomers. This process requires the presence of the histone like HU or 1 HC proteins to facility DNA bending.
b) Dna A protein subunits then successively melt three tandemly repeated 13bp segments in the presence of ATP at >=22*C (open complex).
c) The Dna A protein then guides a Dna B - Dna C complex into the melted region to form a so called prepriming complex. The Dna C is subsequently released. Dna B further unwinds open complex to form prepriming complex.
d) DNA gyrase, single stranded binding protein (SSB), Rep protein and Helicase - II are bound to prepriming complex and now complex is called as priming complex.
e) In the presence of gyrase and SSB, helicases further unwinds the DNA in both directions so as to permit entry of primase and RNA polymerase. Then RNA polymerase forms primer for leading strand synthesis while primase in the form of primosome synthesis primer for lagging strand synthesis.
f) To the above complex, DNA polymerase - III will bind and forms replisome.
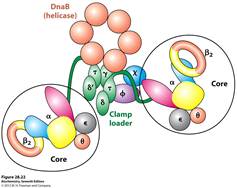
REPLISOME: It is the multiprotein structure that assembles at the bacterial replicating fork to undertake synthesis of DNA. It contains DNA polymerase and other enzymes.
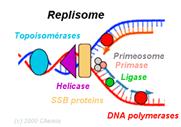
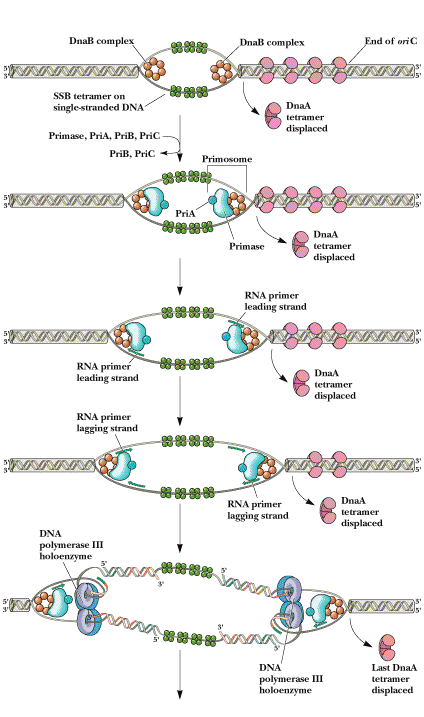
II) ELONGATION:
Now the stage is set for the initiation of synthesis and the elongation to proceed. But this occurs in two mechanistically different pathways in the 5'-->3' template strand and 3'-->5' template strand.
a) Initiation of synthesis and Elongation on the 5'-->3' template (synthesis of leading strand) (If replication fork moves in 3'-->5' direction)
The DNA daughter strand that is synthesized continuously on 5'-->3' template is called leading strand. DNA pol-III synthesizes DNA by adding 5'-P of deoxynucleotide to 3'-OH group of the already presenting fragment. Thus chain grows in 5'-->3' direction. The reaction catalyzed by DNA pol-III is very fast. The enzyme is much more active than DNA pol - I and can add 9000 nucleotides per minute at 37*C. The RNA primer that was initially added by RNA polymerase is degraded by RNase.
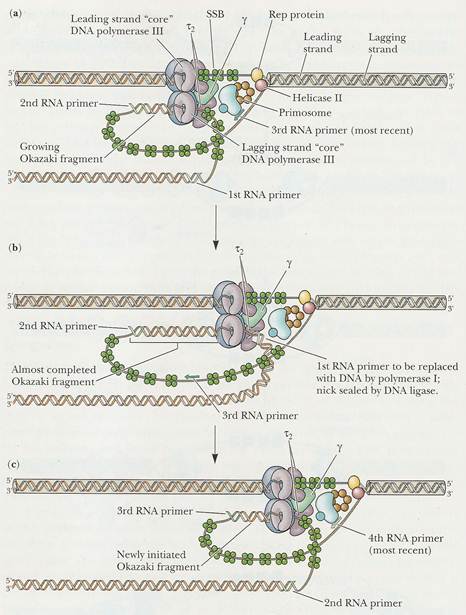
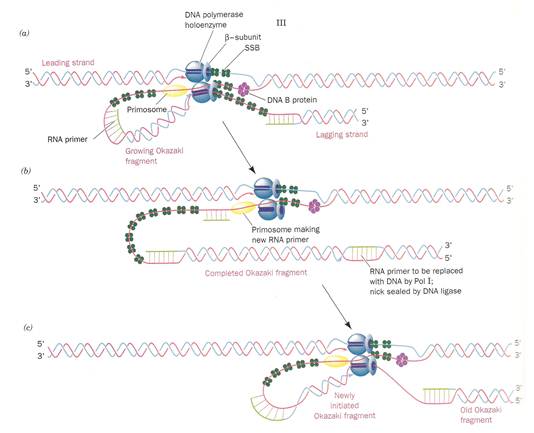
b) Initiation of synthesis and Elongation on 3'-->5' template when fork moves in 3'-->5' direction (Synthesis of lagging strand)
The daughter DNA strand which is synthesized in discontinuous complex fashion on the 3'-->5' template is called lagging strand. It occurs in the following steps:
i) Synthesis of
To the RNA primer synthesized by
primosome, 1000-2000 nucleotides are added by DNA
pol-III to synthesis
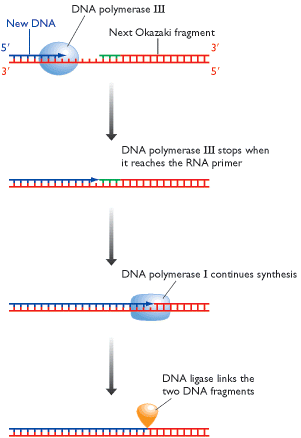
ii) Excision of RNA primer:
When the
iii) Filling the gap (Nick translation)
The gap created by the removal
of primer, is filled up by DNA pol - I using the 3'-OH of nearby
iv) Joining of
Finally, the nick existing between the fragments are sealed by DNA ligase which catalyze the formation of phosphodiester bond between a 3'-OH at the end of one strand and a 5' - phosphate at the other end of another fragment. The enzyme requires NAD for during this reaction.
III) TERMINATION:
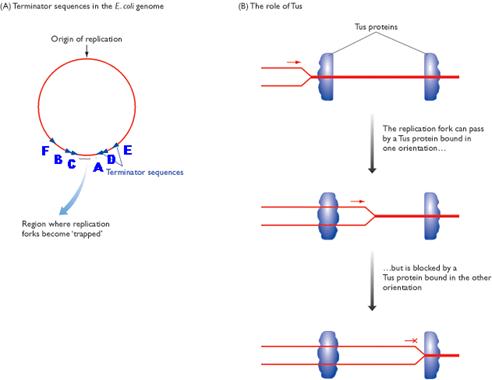
Termination occurs when the two replicating forks meet each other on the opposite side of circular E.Coli DNA. Termination sites like A, B, C, D, E and F are found to present in DNA. Of these sites, Ter A terminates the counter clockwise moving fork while ter C terminates the clockwise moving forks. The other sites are backup sites. Termination at these sites are possible because, at these sites tus protein (Termination utilizing substance) will bound to Dna B protein and inhibits its helicase activity. And Dna B protein released and termination result.
After the complete synthesis, two duplex DNA are found to be catenated (knotted). This catenation removed by the action of topoisomerase. Finally, from single parental duplex DNA, two progeny duplex DNA synthesized.
REGULATION OF PROKARYOTIC REPLICATION:
Especially initiation of replication is regulated. Dna A protein when available in high concentration then ratio of DNA to cell mass is quiet high but at low Dna A concentration, the ratio found to be low. This shows that Dna A protein regulates the initiation of replication.
The sequence most commonly methylated in E.Coli is GATC including in three of 13mer sequence. Thus, the observation that E.Coli defective in the GATC methylation enzyme are very inefficiently replicated, suggests that the DNA replication trigger also responds to the level of OriC methylation.
OTHER MODELS FOR CIRCULAR DNA REPLICATION:
There are two such models are available namely Rolling Circle and D-loop models.
i) Rolling circle model OR s-replication:
In the rolling circle model of replication, a nick is made in one of the strands of the circular DNA, resulting in replication of circle and a tail. This form of replication occurs in the F plasmid or E.Coli Hfr chromosome during conjugation. The F+ or Hfr cell retains the circular daughter while passing the linear tail into the F- cell, where replication of tail takes place. This method is also used in several phages (Viruses), which fill their heads with linear DNA replicated form a circular parent molecule.
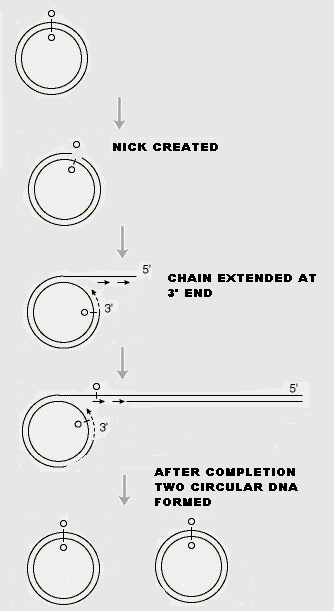
In some cases, because there is no termination point, synthesis often continues beyond a single circle unit, producing concatamers i.e., a series of linked chains, of several circle lengths, which are then processed by recombination to yield normal length circles.
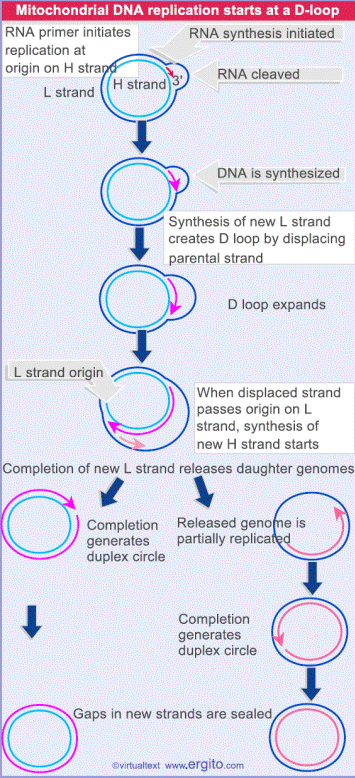
ii) Displacement loop or
D-Loop model:
Chloroplasts and mitochondria in eukaryotic cell have their own circular DNA molecules that appear to replicate by a slightly different mechanism than those described. The origin of replication is at a different point on each of the two parental template strands. Replication begins on one strand, displacing the other while forming a displacement loop or D-loop structure. Replication continues until the process passes the origin of replication on the other strand. The newly synthesized strand was known as leading strand. Replication is then initiated on the second strand in the opposite direction which is the lagging strand. When leading strand completely replicated, only 1/3 of lagging strand is replicated. Then finally the result is two circles.
EUKARYOTIC
DNA REPLICATION (Replication of Linear DNA):
Eukaryotic replication occurs during s-phase of cell cycle. Replication usually occurs only one time in a cell. Replication in eukaryotes occur in five stages namely,
- Pre-initiation
- Initiation
- Elongation
- Termination
- Telomerase function
1. Pre-initiation:
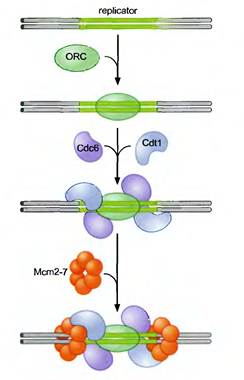
Actually during pre-initiation stage, replicator selection occurs. Replicator selection is the process of identifying the sequences that will direct the initiation of replication and occur in G1 phase. and occurs in Gl (prior to S phase). This process leads to the assembly of a multiprotein complex at each replicator in the genome. Origin activation only occurs after cells enter S phase and triggers the Replicator - associated protein complex to initiate DNA unwinding and DNA polymerase recruitment. Replicator selection is mediated by the formation of pre-replicative complexes (pre-RCs). The first step in the formation of the pre-RC is the recognition of the replicator by the eukaryotic initiator, ORC (Origin recognition Complex). Once ORC is bound, it recruits two helicase loading proteins (Cdc6 and Cdtl). Together, ORC and the loading proteins recruit a protein that is thought to be the eukaryotic replication fork helicase (the Mem 2-7 complex). Formation of the pre-RC does not lead to the immediate unwinding of origin DNA or the recruitment of DNA polymerases. Instead the pre-RCs that are formed during Gl are only activated to initiate replication after cells pass from the Gl to the S phase of the cell cycle.
2. Initiation:
Pre-RCs are activated to initiate replication by two protein kinases namely Cdk (Cyclin Dependant Kinase) and Ddk (Ddt4 Dependant Kinase). Kinases are proteins that covalently attach phosphate groups to target proteins. Each of these kinases is inactive in Gl and is activated only when cells enter S phase. Once activated, these kinases target the pre-RC and other replication proteins. Phosphorylation of these pro-proteins results in the assembly of additional replication proteins at the origin and the initiation of replication.
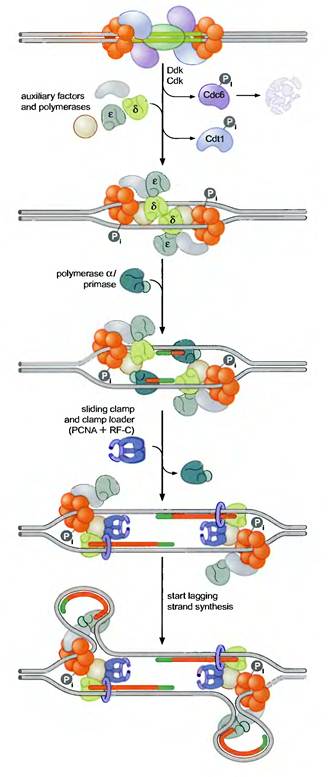
These new proteins include the three eukaryotic DNA polymerases and a number of other proteins required for their recruitment. Interestingly, the polymerases assemble at the origin in a particular order. DNA Pol d and e associate first, followed by DNA Pol a/primase. This order ensures that all three DNA polymerases are present at the origin prior to the synthesis of the first RNA primer (by DNA Pol a/primase). Once present at the origin, DNA Pol a/primase synthesizes an RNA primer and briefly extends it. Thus initiation of replication started.
3. Elongation:
The resulting primer-template junction is recognized by the eukaryotic sliding clamp loader (RF-C), which assembles a sliding clamp (PCNA) at these sites. Either DNA Pol d or e recognizes this primer and begins leading strand synthesis. After a period of DNA unwinding, DNA Pol a/primase synthesizes additional primers, which allow the initiation of lagging strand DNA synthesis by either DNA Pol d or e. In the diagram, Pol d was used for leading strand and Pol e was used for lagging strand synthesis. DNA Pol e possess activity to remove primer and fills the gap with DNA like DNA Pol I in prokaryotes. SSB like activity was played by replication protein A (RP A) which is denoted as accessory factors during replication.
4. Termination:
When the replication forks meet each other, then termination occurs. It will result in the formation of two duplex DNA. Eventhough replication terminated, 5’ end of telomeric part of the newly synthesized DNA found to have shorter DNA strand than the template parent strand. This shortage corrected by the action of telomerase enzyme and then only the actual replication completed.
5. Terlomerase Function:

In Linear eukaryotic chromosome, once the first primer on each strand is remove, then it appears that there is no way to fill in the gaps, since DNA cannot be extended in the 3'-->5' direction and there is no 3' end upstream available as there would be in a circle DNA. If this were actually the situation, the DNA strand would get shorter every time they replicated and genes would be lost forever.
Elizabeth Blackburn and her colleagues have provided the answer to fill up the gaps with the help of enzyme telomerase. So, that the genes at the ends, are conserved. Telomerase is a ribonucleoprotein (RNP) i.e. it has RNA with repetitive sequence. Repetitive sequence varies depending upon the species example Tetrahymena thermophilia RNA has AACCCC sequence and in Oxytrica it has AAAACCCC. Telomerase otherwise known as modified Reverse Transcriptase. In human, the RNA template contains AAUCCC repeats. This enzyme was also known as telomere terminal transferase.
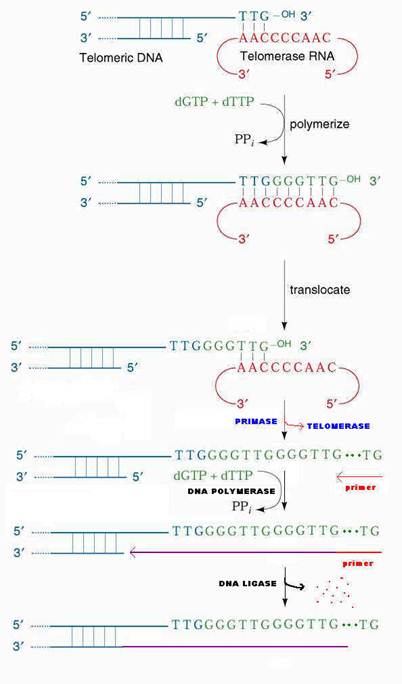
The 3'-end of the lagging strand template basepairs with a unique region of the telomerase associated RNA. Hybridization facilitated by the match between the sequence at the 3'-end of telomere and the sequence at the 3'-end of the RNA. The telomerase catalytic site then adds deoxy ribonucleotides using RNA molecule as a template, this reverse transcription proceeds to position 35 of the RNA template. Telomerase then translocates to the new 3'-end by pairing with RNA template and it continues reverse transcription. When the G-rich strand sufficiently long, Primase can make an RNA primer, complementary to the 3'-end of the telomere's G-rich strand. DNA polymerase uses the newly made primer to prime synthesis of DNA to fill in the remaining gap on the progeny DNA. The primer is removed and the nick between fragments sealed by DNA ligase.
REGULATION OF EUKARYOTIC REPLICATION:
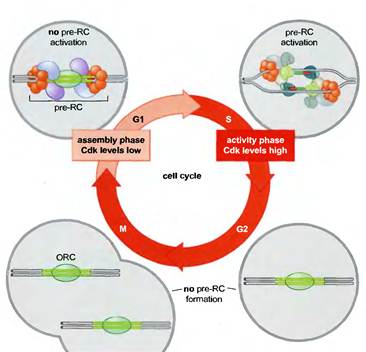
The tight connection between pre-RC function, Cdk levels, and the cell cycle ensures that the eukaryotic genome is replicated only once per cell cycle (Figure 8-32). Active Cdk is absent during Gl, whereas elevated levels of Cdk are present during the remainder of the cell cycle (S, G2, and M phases). Thus, during each cell cycle there is only one opportunity for pre-RCs to form during Gl and only one opportunity for those pre-RCs to be activated during S, G2, and M phase, although in practice all pre-RCs are activated or disrupted by replication forks in S phase. Pre-RCs are disassembled after they are activated or after the DNA to which they are bound is replicated. These exposed replicators are then available for new pre-RC formation and rapidly bind to ORC. Despite the presence of the initiator at these sites, the elevated levels of Cdk activity in S, G2, and M phase cells prevents the association of the other members of the pre-RC complex with ORC. It is only when cells segregate their chromosomes and complete cell division that Cdk activity is eliminated and new pre-RC complexes can form.
Inhibition of replication
was achieved by antisense RNA. Once antisense RNA produced for particular
gene, it bound with single strand DNA in open complex and inhibits the movement
of replisome and thus replication. Antisense RNA synthesized in opposite
direction to the normal RNA synthesis.
INHIBITORS OF REPLICATION:
Dauromycin and Adriamycin:
They are synthetic chemotherapeutic agents and are inhibitors of both DNA replication and transcription in prokaryotes. These are presumably act by interfering with the passage of both DNA and RNA polymerase. They have planar aromatic ring system which gets intercalated between GC pairs of the double helical structure. Thus, they prevent its replication and transcription. Adriamycin is otherwise known as doxorubicin. Doxorubicin is a cytotoxic anthracycline antibiotic isolated from cultures of Streptomyces peucetius var. caesius. Doxorubicin consists of a naphthacenequinone nucleus linked through a glycosidic bond at ring atom 7 to an amino sugar, daunosamine. Doxorubicin binds to nucleic acids, presumably by specific intercalation of the planar anthracycline nucleus with the DNA double helix. The anthracycline ring is lipophilic, but the saturated end of the ring system contains abundant hydroxyl groups adjacent to the amino sugar, producing a hydrophilic center. The molecule is amphoteric, containing acidic functions in the ring phenolic groups and a basic function in the sugar amino group. It binds to cell membranes as well as plasma proteins.
Actinomycin - D
It is an antibiotic produced by streptomyces and inhibits replication and transcription. It acts by intercalating its phenoxazone ring between two successive GC pairs in duplex DNA. Actinomycin D has two identical pentapeptides which have unusual composition of D-Valine and Sarcosine which stabilizes this intercalating interaction. It was the first antibiotic shown to have anti-cancer activity, but is not normally used as such, as it is highly toxic, causing damage to genetic material. Actinomycin-D is marketed under the trade name Dactinomycin. Actinomycin-D is one of the older chemotherapy drugs which has been used in therapy for many years. It is a clear, yellow liquid which is administered intravenously and most commonly used in treatment of a variety of cancers.
Ethidium bromide and proflavin
They inhibit both replication and transcription by intercalation. As with most fluorescent compounds, it is aromatic. The main portion of the molecule is a tricyclic structure with aniline (aminobenzene) groups on either side of a pyridine (a six-atom, nitrogen-containing, aromatic ring). The dibenzopyridine structure is known as a phenanthridine. The reason for ethidium bromide's intense fluorescence after binding with DNA is probably not due to rigid stabilization of the phenyl moiety, because the phenyl ring has been shown to project outside the intercalated bases. In fact, the phenyl group is found to be almost perpendicular to the plane of the ring system, as it rotates about its single bond to find a position where it will abut the ring system minimally. Instead, the hydrophobic environment found between the base pairs is believed to be responsible. By moving into this hydrophobic environment and away from the solvent, the ethidium cation is forced to shed any water molecules that were associated with it. As water is a highly efficient fluorescent quencher, the removal of these water molecules allows the ethidium to fluoresce. This property is used to identify the presence of DNA in gel und UV light.
Novobiocin and oxolinic Acid
Prokaryotic DNA gyrases are
specifically inhibited by two classes of antibiotics. One of these classes includes the
streptomyces-derived novobiocin and the other contains the clinically useful
synthetic antibacterial agent oxolinic acid. Both classes of antibiotics profoundly
inhibit bacterial DNA replication and transcription, thereby demonstrating the
importance of properly supercoiled DNA in these processes. Studies using antibiotic-resistant E.Coli mutants have demonstrated
that oxolinic acid associates with DNA gyrase’s A
subunit and novobiocin binds to its B subunit.
When DNA gyrase is incubated with DNA and oxolinic
acid, and subsequently denatured with guanidinium chloride, it’s A subunits
remain covalently linked to the 5’-ends of both cut strands through phosphotyrosine linkages.
Apparently oxolinic acid interferes with
gyrase action by blocking the strand breaking-rejoining process. Novobiocin, on the other hand, prevents ATP
from binding to the enzyme.
Aphidicolin
Aphidicolin inhibits DNA pol a of eukaryotes, so it
inhibits eukaryotic replication.
Aphidicolin also inhibits DNA pol d and DNA pol e.
Thus Aphidicolin
inhibits both leading and lagging strand synthesis in eukaryotes.
Rifamycin
Rifamycin inhibits RNA
polymerase, so RNA primer for leading strand synthesis is not available. Thus replication inhibited.
The rifamycins
are a group of antibiotics which are synthesized either naturally by the
bacterium Amycolatopsis mediterranei,
or artificially. The rifamycin group includes the "classic" rifamycin
drugs as well as the rifamycin derivatives Rifampicin, Rifabutin and Rifapentine.
The biological activity of rifamycins relies on the
inhibition of DNA-dependent RNA synthesis. This is due to the high affinity of rifamycins to prokaryotic RNA polymerase.
*******