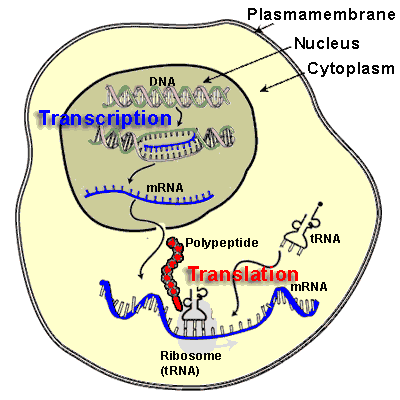
TRANSCRIPTION
The DNA strand that is transcribed for a given mRNA is termed the template strand or Non-coding strand or Non-sense strand. Its complementary DNA strand will be referred to as the Non-template strand or coding strand or sense strand. The transcribed RNA has the sequence of non-template strand with the exception of U instead of T.
DNA DIRECTED RNA POLYMERASE:
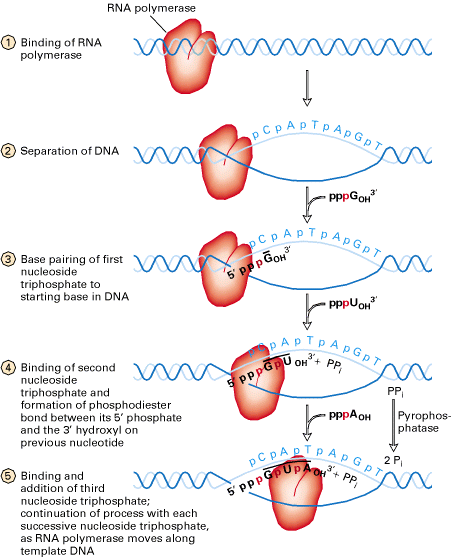
The enzyme DNA directed RNA polymerase begins the synthesis of RNA and adds ribonucleotides to the 3-end of RNA in which DNA molecules template strand determines the base sequence of RNA.
The ribonucleotide incorporation reaction requires energy provided by triphosphates of the four nucleotides. This reaction is driven by the hydrolysis of the pyrophosphate released upon incorporation of each nucleotide monophosphate into the growing RNA chain. The reactions are as follows:
(RNA)n + NTP -------------ΰ (RNA)n+1 + PPi
PPi --------ΰ 2Pi
Where
NTP ΰ Any triphosphates nucleotide
Pi ΰ Inorganic phosphate
PPi ΰ Pyrophosphate
n ΰ Number of nucleotide in RNA
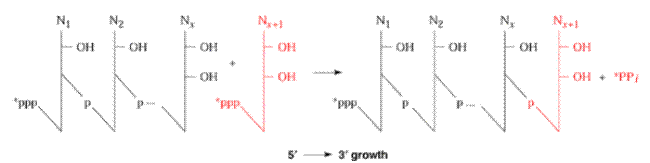
The second reaction requires the enzyme inorganic pyrophosphatase to split the pyrophosphate, releasing the energy that drives the incorporation reaction. During RNA polymerase function two things occur simultaneously. First, DNA is being decoded. That is, the polymerase enzyme reads the deoxynucleotides of the anticoding or template, strand of DNA and base pairs those nucleotides with the appropriate ribonucleotides. Second, a phosphodiester bond is produced between the 3-position of the last ribonucleotide of the m-RNA and the 5-position of the new ribonucleotide to be incorporated. So, decoding of the gene and m-RNA synthesis occurs at the same time.
Mechanism:
Conditions for RNA Polymerase Activity (Pre-requisite):
The following are the requirements for DNA directed RNA polymerase functions
a) A template of a double stranded DNA
b) All four ribonucleotides
c) Mg2+ ions.
Unlike DNA polymerase, RNA Polymerase does not require primer.
Prokaryotic RNA Polymerase:

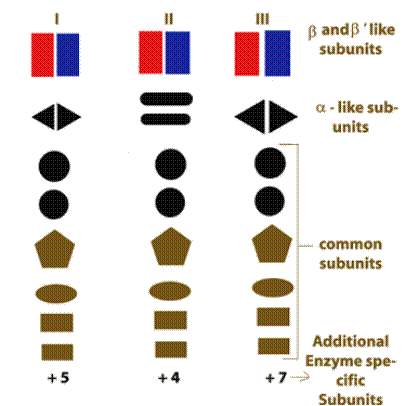
E.Coli RNA
polymerase is a complex enzyme consisting of six subunits a2, b, b,w and s. The beta subunit (b) has a molecular weight of
150,000, Beta prime (b) 160,000, alpha (a) 40,000, omega (w) 11,000 and sigma (s) 70,000. The complex RNA
polymerase enzyme of E.Coli,
the holoenzyme, is composed of a core enzyme and a sigma factor. The core enzyme is composed of five subunits a2, b, b and w. Core enzyme can continue transcription after
initiation but holoenzyme is necessary for correct initiation of transcription.
|
S.No |
Factors |
Functions |
|
1. |
s |
Recognition of Promoter sequence and aids the proper binding of RNA Polymerase to DNA initiation site. |
|
2. |
b |
Binding of RNA Polymerase to template |
|
3. |
b |
Polymerization function |
|
4. |
a, w |
Structural component of RNA Polymerase |
Eukaryotic DNA directed RNA polymerase:
RNA Polymerase I :
RNA Polymerase II:
It has been the most intensively studies of the three polymerases. The enzyme is composed of two large polypeptides and from 6 to 8 smaller polypeptides. This polymerase recognizes three different elements of a gene.
a) A selector sequence containing a TATA box and a short sequence.
b) An upstream promoter sequence
c) An enhancer sequence which may be located at different site in different genes.
Seven transcription factors required by RNA Pol-II for specific binding of the enzyme to the DNA promoter and to initiate transcription.
RNA Polymerase III :
Template Independent RNA Polymerases
:
a) t-RNA specific Nucleotidyl transferase:
It adds CCA sequence to 3-end of t-RNA during post transcriptional modification of pre-t-RNA.
b) Poly (A) Polymerase :
It adds poly A tail to 3-end of hn-RNA during post transcriptional modification of eukaryotic pre-m-RNA.
TRANSCRIPTION OF PROKARYOTIC RNA:
Transcription involves three stages:
1) Initiation
2) Elongation
3) Termination
1) Initiation:
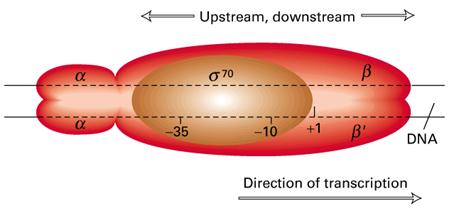
Transcription begins when the DNA directed RNA Polymerase associates with the sigma factor to produce holoenzyme. The sigma factors allows the polymerase to bind specifically at the genes promoter sequence. There are two important promoter regions present. They are Pribnow box and -35 sequence.
Pribnow box (-10 region):
-35 sequence:
It is present 16-19 bases upstream from the Pribnow box. It is a hexameric consensus sequence TTGACA. This region is the initial binding site for the s subunit of RNA polymerase.

The first stage of transcription is the formation of an open promoter complex. First, the s subunit of the RNA polymerase recognizes and binds to -35 sequence followed by the binding of core enzyme.
Then the RNA Polymerase by virtue ofo its large size spans region of 17 to 19 base pairs and gets bound to the Pribnow box. This early stage structure of binding of RNA Polymerase (Holoenzyme) to the -10 region is called closed promoter complex.
RNA Polymerase inturn cause localized melting i.e. unwinding of DNA helix. This conformation stage is called open promoter complex. Unwinding spans in a region of about 10 basepairs from left end of the Pribnow box and extending about 20 base pairs after the position of the first transcribed base. The melting is necessary for the pairing of incoming ribonucleotides.
Once RNA Polymerase moves on, the DNA behind the enzyme closes as the hydrogen bonds of the DNA base pairs reforms. The enzyme reads the template strand in a 3 to 5 direction as it synthesizes m-RNA in a 5ΰ3 direction.
2) Elongation:
After several nucleotides (mostly eight) are added to the growing chain, RNA Polymerase undergoes a conformational change and the s subunit dissociates. Therefore, the chain elongation process is carried out by the core enzyme. Core enzyme continues reading the template strand and joining ribonucleotides by addition to the 3-end of the growing chain synthesis of m-RNA is therefore in the 5ΰ3 direction as the template is decoded in 3ΰ5 direction. The strands of double stranded nucleic acids, even in temporary hybrids such as DNA-RNA molecules, must be antiparallel if hydrogen bonding across the strand is to takes place.
The energy required for synthesis is provided by the triphosphates ribonucleosides. These ribonucleotides are the energy sources, building blocks and the information components for m-RNA synthesis.
3) Termination:
The last stage in m-RNA synthesis is chain growth termination. Synthesis of m-RNA is ended by any one of the following ways namely
a)
b)
The DNA sequences, often referred to as transcription terminators are either rho-dependent or rho-independednt. In either case, a so called stem-loop or hairpin structure, is formed. RNA synthesis terminates shortly after this structure is formed.
The stem-loop forms at the 3-end of the m-RNA, because at the 5-end of the template DNA an unusual sequence of nucleotides occurs. This sequence is known as dyad symmetry i.e. inverted base sequence with central repeat sequence. That is, read in a 5ΰ3 direction, the DNA nucleotide sequences of the two strands are identical. For example,
5 GGCTCCTTTTGGAGCC 3
3 CCGAGGAAACCTCGG 5
When m-RNA is transcribed from the template strand (3ΰ5), the resulting sequence is
5 GGCUCCUUUUGGAGCC 3

The molecule is self complementary, so a hairpin or stem loop can form.
A) Rho-independent Termination:
In Rho-independent termination, the template inverted repeat of DNA is followed by a series of adenines. This series produces a run of perhaps half a dozen Uracils in the m-RNA. So, the m-RNA in rho-independent termination has the following structure.
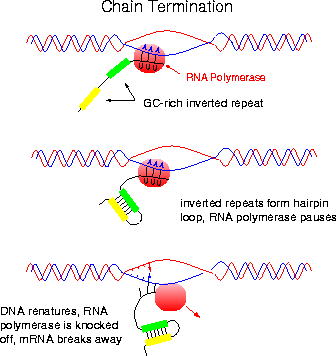
At the point where the poly U sequence is attached to the DNA sequence, the hybrid DNA-RNA is unusually weak (A-U bonds are weak) and it requires very little energy to break the hydrogen bonds holding the two strands together. When separation occurs, m-RNA synthesis, transcription stops. This type of termination is rho-independent; no termination factor is required.
B)
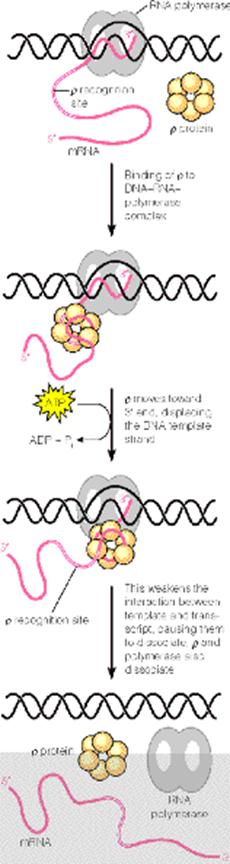
It is proposed that first rho protein binds to the 5-end of a nascent RNA chain and then moves along the RNA, using the hydrolysis of ATPs to provide the necessary energy. Then, when RNA Polymerase pauses at certain sites having high GC sequences or stem-loop structure it catches up and binds with RNA Polymerase. When rho binds with RNA Polymerase it assume ATPase and brings about the hydrolysis of RNA chain after which the rho factor and the enzyme dissociates from the template.
It also appears that termination is not absolutely rho dependent or rho-independent. Rather, rho-independent termination can utilize rho and rho dependent termination can proceed in the absence of the protein.
Regulation of Prokaryotic
Transcription:
(Control of Prokaryotic Transcription)
There are different ways in which transcription of Prokaryotes controlled namely
a) Promoters activity
b) Repressors activity
c) Catabolite Repression
d) Dual Positive and Negative control
e) Attenuation
f) Stringent Response
a) Promoters Activity:
There are different types of promoter sequences present in DNA molecules. Depending upon the conservity of the sequences match with Pribnow box, promoters classified into two different types. They are strong promoters and weak promoters. Strong Promoters have the sequence which resemble almost similar to Pribnow box whereas weak promoters sequence varied to larger extent. There are different types of s - molecules which intern has different extent of affinity to promoter sequences. So strong promoters sequence with high affinity s - molecules will provide increased transcription rate and vice versa.
b) Repressor Activity:
Repressors will bind to the operator region. So, that RNA Polymerase is unable to bind to the promoter region because promoter and operators regions are overlapping to each other. Because of this, there is no expression of genes example Lac Operon. At low lactose concentration, repressors prevent the expression of Lac Operon.
c) Catabolite Repression:
Glucose is E.Colis metabolite of choice; the availability of adequate amounts of glucose prevents the full expression of genes specifying proteins involved in the fermentation of numerous other catabolites including lactose, arabinose and galactose, even when they are present in high concentrations. This phenomenon, which is known as catabolite repression, prevents the wasteful duplication of energy producing enzyme systems.
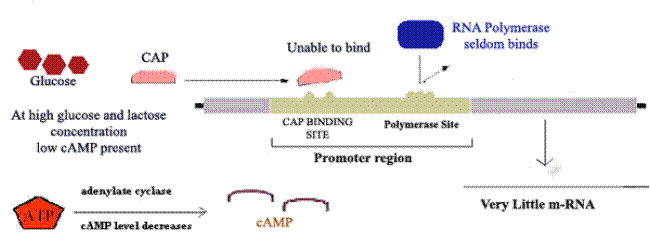
Ex: Lac Operon
When glucose
concentration increased inside cell then concentration of cAMP decreased which
intern decrease
d) Dual Positive and Negative Control:
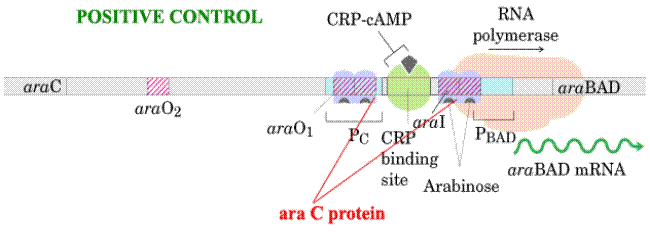
Example: Ara Operon
Ara C protein plays dual positive and negative control on Ara operon.
In the presence of arabinose, the Ara C protein binds to the ara I region and when bound to cAMP, the CAP protein binds to a site adjacent to ara I. This binding stimulates the transcription of the structural genes.
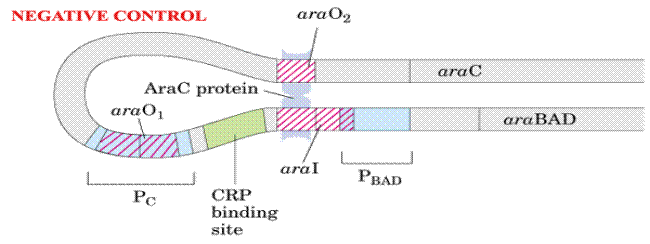
In the absence of arabinose, the Ara C protein binds to both ara I and ara O regions, forming a DNA loop. This binding prevents transcription of the ara operon.
e) Attenuation:
Attenuation is the process for regulation of prokaryotic gene expression. It occurs in anabolic in anabolic operons i.e. the gene products are responsible for the synthesis of some compounds mainly aminoacids like tryptophan, Histidine, Leucine etc., these operons have a sequence known as Leader sequence inbetween operator and structural genes. This sequence contains four segments. These segments have the sequence such that they can form stem and loop structure. When 3rd and 4th segments forms stem and loop structure, termination of transcription would occur. But when 2nd and 3rd segment forms stem and loop structure, no transcriptional termination and transcription continues.
In prokaryotes, translation is closely coupled with transcription i.e. during transcription after small segment of gene transcribed; ribosome attached to it and initiates translation.
Example: Trp Operon
Segment 1 of transcribed m-RNA of Trp Operon contain codons for Tryptophan which intern regulate transcription. When tryptophan is abundant, segment 1 of the trp m-RNA is fully translated. Segment 2 enters the ribosome which enables Segment 3 and 4 to base pair. This base paired region signals RNA Polymerase to terminate transcription.
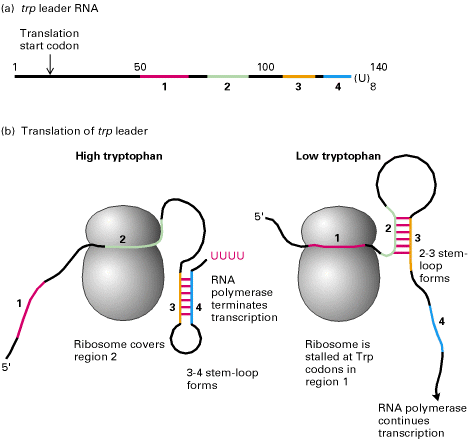
When tryptophan is scarce, the ribosome stalled at the codon of segment 1. Segment 2 interacts with segment 3 instead of being drawn into the ribosome and so segments 3 and 4 cannot pair. Consequently, transcription continues.Thus the 2-3 segments stem and loop structure known as attenuator. This process is known as attenuation.
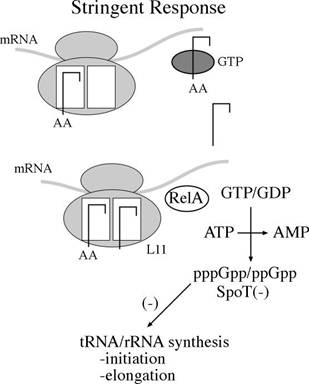
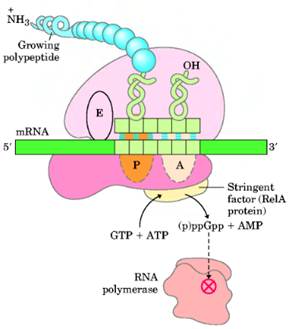
f) Stringent Response:
Stringent Response controls transcription of r-RNA and t-RNA. Guanosine tetra phosphate [ppGpp] is the prime component for stringent response. When charged t-RNA not available for codons, uncharged t-RNA attached to A Site of ribosome. So that ribosome stalls at that site. This favors synthesis of ppGpp by rel A. ppGpp then binds with RNA Polymerase and inhibit its action towards promoters for r-RNA and t-RNA genes. But transcription of genes for amino acid synthesis, lac operon and ara operon are initiated. When charged t-RNA [aminoacyl t-RNA] available, then the level of ppGpp decreased by the action of spot. Once ppGpp concentration decreased, inhibition of RNA Polymerase released and thus transcription of r-RNA and t-RNA occurs.
EUKARYOTICE TRANSCRIPTION:
In eukaryotes, different RNA Polymerases involved in transcription of different type of RNAs. Hence mechanism differs. Transcription by each RNA Polymerases studied separately. They are
a) Transcription by RNA Pol I
b) Transcription by RNA Pol II
c) Transcription by RNA Pol III
A) TRANSCRIPTION BY RNA POLYMERASE - I:
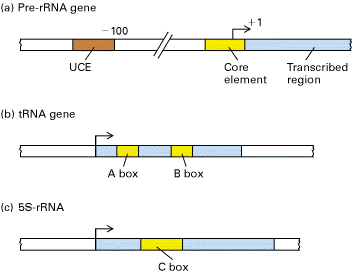
RNA Pol I dedicated to the synthesis of only one type of RNA molecule, called pre-r-RNA. The primary pre-r-RNA transcript is processed into the 18S, 5.8S, and 28S rRNAs found in vertebrate ribosomes or their functional equivalents in other eukaryotes.
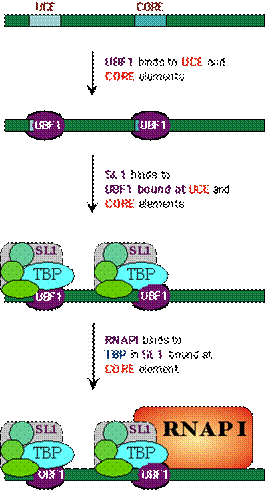
The control region of pre-RNA transcription units contains a core promoter element which overlaps the start site and an upstream control element (UCE) located ~100 base pairs upstream. Upstream binding factor (UBF) binds to the UCE and core element and the two bound molecules are through to make protein-protein interactions causing the intervening DNA to loop out. Selectivity factor (SL1) then binds to the UBF DNA complex and the remaining free segment of the core element. SL1 is a multimeric protein composed of TBP and three TBP-associated factors with Mws of 110, 63 and 48kDa. It is also species specific factor. Finally, RNA Polymerase I binds, completing assembly of the initiation complex.
After the formation of initiation complex, Pol-I unwind DNA and initiates transcription. Transcription is then elongated with RNA Pol I alone. Finally, transcription is terminated in a rho dependent termination manner.
B) TRANSCRIPTION BY RNA POLYMERASE II :
Promoters for RNA Pol II
1) TATA box [Goldberg Hogness box] [-25 region]:
It is located about 25 base pairs upstream from the transcription start site and has a consensus sequence TATAAAT. The TATA box is usually flanked by high GC sequences.
2) GC box:
It is located about 40 basepairs upstream from transcription start site. It has consensus sequence GGGCGG.
3) CAAT box:
It is located about 75 base pairs upstream from the transcription start site and has a consensus sequence GGT/CCAATCT.
4) Upstream Activating Sequence:
These sequence are also known as hypersensitive sites and are thought to influence many m-RNA synthesis for many specific proteins.

Transcription by RNA Pol II occurs in four stages. They are
I Formation of Initiation Complex
II Initiation
III Elongation
IV Termination
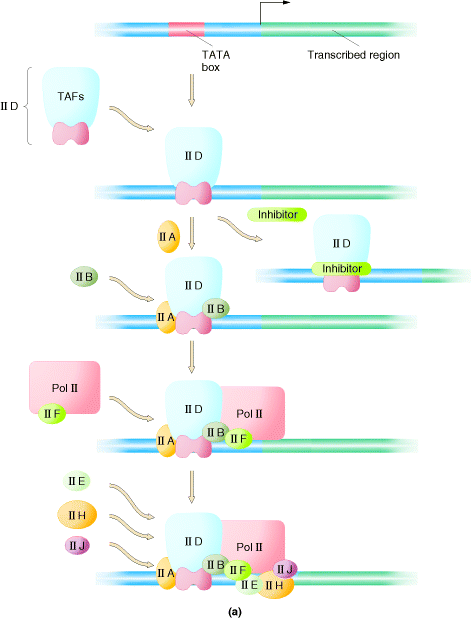
I) Formation of Initiation Complex:
Initiation complex begins with the binding of transcription factor TF II D to the TATA box. TF II D is composed of one TATA box binding subunit called TBP and more than either other subunits (TAFs), represented by one large symbol. TF II A binds to TF II D promoter complex to form DA complex. TF II B then binds to D A complex, followed by binding of a preformed complex between TF II F and RNA Polymerase II. Finally, TF II E, TF II H and TF II J must add to the complex, in that order, for transcription to be initiated. Initiation by RNA Pol II requires hydrolysis of the b - g bonds of ATPs. One of the last factors to add to the complex, TF II H, which has DNA helicase activity, can use the energy from hydrolysis of ATP to separate the strands of the duplex template DNA. This protein is suspected to mediate unwinding of the strands at the start site allowing the Polymerase to initiate transcription. TF II H also has a protein kinase activity, which can transfer the g - phosphates of ATPs to multiple serines in C-terminal repeat domain (CTD) of the largest RNA Pol II subunit.
II ) Initiation:
After open promoter complex formation in which the template strand is exposed for transcription, initiation occurs. RNA Pol II contain two nucleotide binding sites, called the initiation site and elongation site. The initiating nucleoside triphosphates bind to the enzyme and forms hydrogen bond with a complementary base in DNA at initiation site. Elongation site is then filled with nucleoside triphosphates that are selected by its ability to hydrogen bonded to the next base in the DNA strand. The two nucleotides are then joined together through phosphodiester bond and the first base is released from the initiation site. Then RNA Polymerase moves in the relative direction so that enzyme shifted exactly by one nucleotide distance. After initiation TF II E released from initiation complex.
III) Elongation:
After few nucleotides added to the growing pre-mRNA chain, TF II H adds phosphate to serine residues in CTP of Pol II. This phosphorylation releases attachment of CTD to TF IID. After phosphorylation, RNA Polymerase II can move freely on DNA template and TF II H dissociates. Therefore, the chain elongation process is carried out by the RNA Pol-II and TF II J and F. RNA Pol-II unwinds DNA continuously as the enzyme extends the growing RNA chain. Nascent RNA chain grew in 5ΰ3 direction.
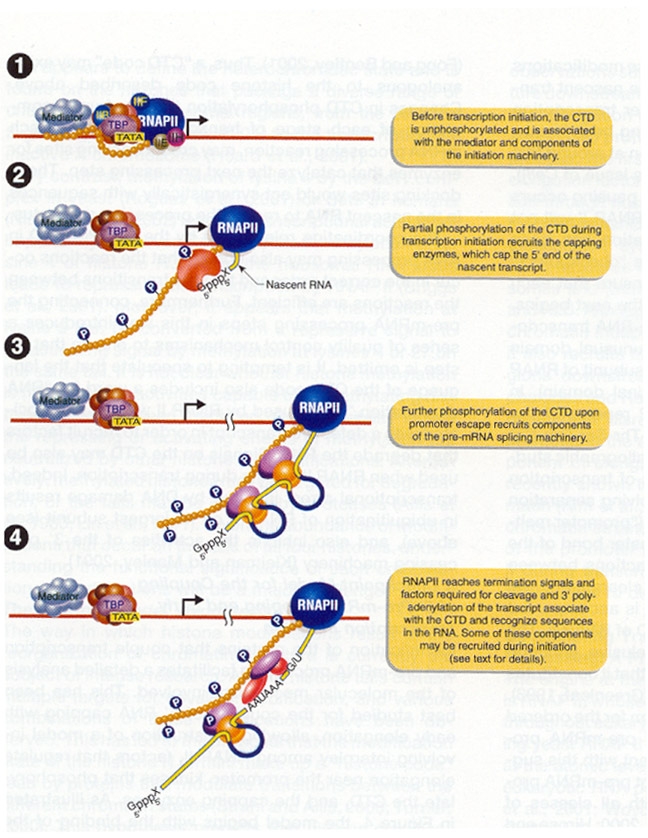
IV) Termination:
Termination does not require rho factor. Termination is rather carried out by the core enzyme itself by virtue of its ability to recognize certain sites in the DNA template which are called termination signal sites. This has three distinguishing features. They being
a) It has a inverted repeat base sequence containing central repeat sequence which allows the formation of stem and loop configuration leading to the excision of the RNA transcript.
b) It has high GC regions
c) It has high AT regions
All the above mentioned structural features of the termination site helps in the formation of hairpin structure in growing RNA chain which gets easily dissociated from DNA template and hence its termination.
C) TRANSCRIPTION BY RNA POL III:
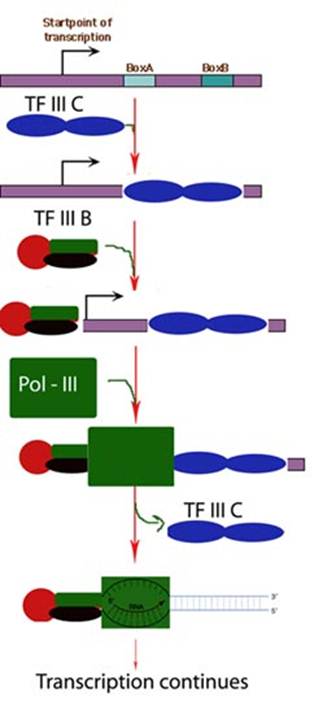
i) Transcription of t-RNA
genes:
First, TF II C, a large multisubunit protein, binds with high affinity to the B box promoter and with low affinity to the A box. TF III C acts as an assembly factor for binding the trimeric TF III B to any DNA sequence upstream of the t-RNA gene. TF III B is made up of three subunits. One is TBP. The second called BRF (TF II B related factor) is similar in sequence to TF II B. The third subunit of TF III B is a 90 kD polypeptide called B. Once TF III B binds, then RNA Polymerase can bind and TF III C is released. RNA Polymerase then initiates transcription in the presence of ribonucleoside triphosphates. Enzyme does not require hydrolysis of an ATP b - g bond similar to RNA Pol I. After initiation, RNA Polymerase elongates transcription and finally terminates transcription in a rho independent termination mechanism.
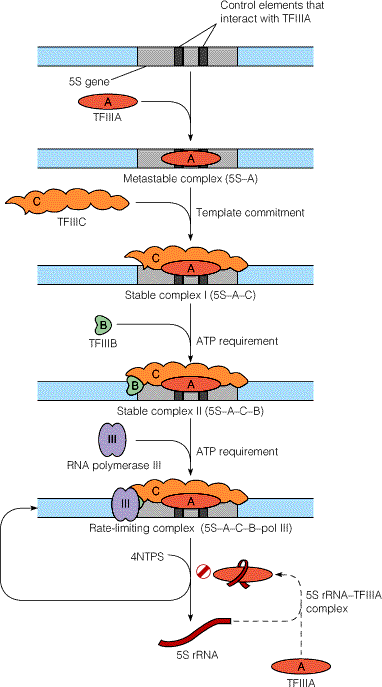
ii) Transcription of 5S r-RNA gene:
Synthesis of 5S
r-RNA is initiated by binding of TF III A to the C box. Once TF III A has bound, TF III C binds to
the gene at a similar position relative to the start site as when TF III C
binds to a t-RNA gene. TF III B then
binds, interacting analogously with TF III C as it does in a t-RNA gene. Once TF III B has bound, RNA pool III and
binds and initiates transcription. TF
III A thus acts an assembly factor for binding of TF III C; TF III C then act
as an assembly factor for TF III B. RNA
Pol III elongates transcription and terminate it in a rho dependent
termination mechanism.
REGULATION OF EUKARYOTIC
TRANSCRIPTION:
Eukaryotic transcription regulated with the help of any one of the following components, namely,
i) Cis-acting elements
ii) Trans-acting elements (Transcriptional factors)
iii) Hormones
iv) Antiterminants
i) Cis-acting elements:
Cis-acting elements are specific sequence in DNA. They are of different types like promoter, proximal promoter element, enhancer and silencers. When specific components bind to promoter, promoter proximal element and enhancer, transcription initiated and rate increased whereas when it bound to silencer, transcription suppressed.
ii) Trans-acting elements:
Transcriptional factors like TF IIB, SL1, TFIIIB etc., are transacting elements. When transcriptional factors are present along with polymerase then transcription occurs. When their concentration decreased, transcriptional rate also decreased.
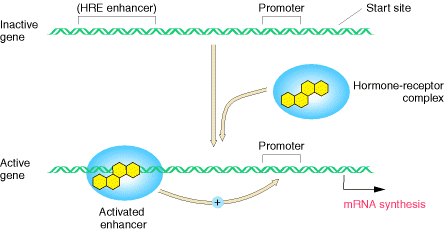
iii) Hormones Steroid Hormones:
Steroid Hormones enters into the cell and it binds with receptor to form hormone receptor complex. Then HR complex translocates inside the nucleus. Within the nucleus, HR binds with hormone response element which intern stimulates transcription. Absence of these hormones decrease transcription rate.
iv) Antiterminators:
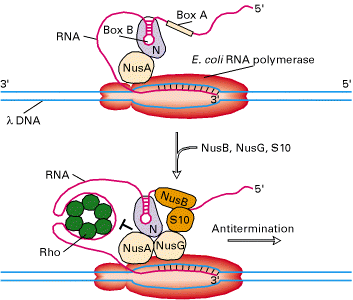
In some genes, termination signal sequence present even after promoter itself. Depending upon the need of specific gene products, this termination signal may cause pretermination. In some cases, this premature termination was prevented by certain factors. They are referred as Antiterminators.
E.g Expression of C-myc gene
Presence of growth factors includes expression of C-myc gene which intern prevents the premature termination. So growth factor act as Antiterminators.
POST TRANSCRIPTIONAL MODIFICATION OR PROCESSING:
The immediate products of transcription, the primary transcripts, are not necessarily functional entities. In order to acquire biological activity, many of them must be altered in several ways.
i) by the exo and endo nucleolytic removal of polynucleotide segments
ii) by appending nucleotide sequence to their 3 end and 5-ends and
iii) by the modification of specific nucleosides
The three major classes of RNAs mRNA, rRNA and tRNA, are altered in different ways in prokaryotes and eukaryotes. These modifications after transcription are referred as Post transcriptional modifications or processing.
m-RNA PROCESSING:
Prokaryotic m-RNA:
In prokaryotes, most primary m-RNA transcript functions in translation without further modification. Ribosomes in prokaryotes usually commence translation on nascent m-RNAs itself. So, prokaryotic m-RNA does not undergo post transcriptional processing.
Eukaryotic m-RNA processing:
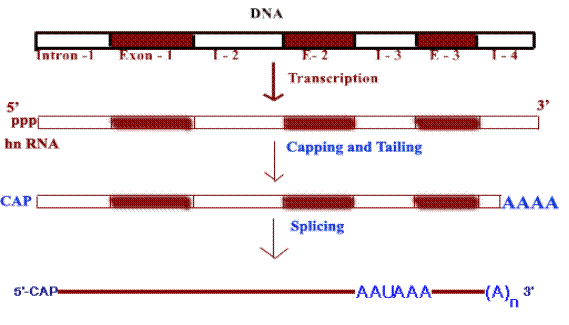
In eukaryotes m-RNAs are synthesized in nucleus while translation occurs in the cytosol. Apart from spatial segregation there also exists a finite temporal lag i.e. to say the transcription and translation does not go hand by hand. Infact, m-RNA synthesized as heterogeneous nuclear RNA [hnRNA] or pre-m-RNA which is the primary m-RNA transcript in the nucleus. It thereafter undergoes extensive post transcriptional processing while still in the nucleus, to form mature m-RNA which then gets transported to cytosol to get associated with ribosomes for the translation process to commence. Processing of m-RNA involves the following stages,
a) Capping
b) Tailing
c) Splicing
d) Methylation
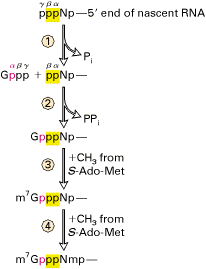
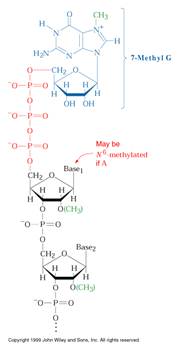
a) Capping:
All eukaryotic m-RNAs have a cap structure at the 5end consisting of a 7-methyl guanosine residue join to the transcript via 5-5 triphosphates bridge. The cap structure is attached to the 5-end of the growing transcript by guanylyl transferase before it is greater than 20 nucleotides long. There are three types of capping
Cap O:
When the two leading nucleosides are not methylated at 2 position, are called cap O type which occurs predominantly in unicellular eukaryotes.
Cap 1:
When the first nucleoside following the 7-mehtyl guanosine methylated at the 2position, it is called cap 1 structure. Such capping occurs in most of the multicellular organisms.
CAP 2:
When the first two nucleosides following 7-methyl guanosine is methylated at 2 position, it is called cap2 structure and it is found in some eukaryotes.
Significance of capping:
I) Enhancement of translation ability of m-RNA. Recent studies showed that capping of m-RNA is essential for binding to the smaller subunit of ribosomes.
II) Capping protects m-RNA from ribonuclease (RNase)
CAP STRUCTURE:
b) Tailing:
Tailing is a process in which poly A tail with around 200 adenosine residues attached to 3-end of hnRNA. Events in Tailing shown in the following diagram.
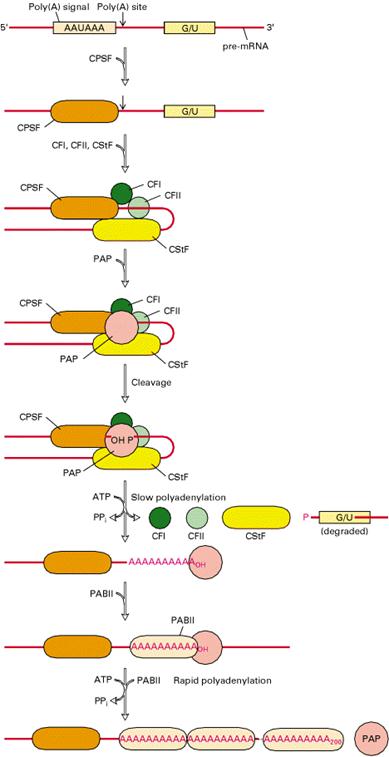
Significance:
Experimental evidences are shown that poly A stabilizes m-RNA. m-RNAs which have poly A tail have greater life time in cytosol whereas other m-RNAs which have no poly A tail have lifetime less than 30 minutes in cytosol.
c) Splicing:
The most striking differences between eukaryotic and prokaryotic structural genes are that the coding sequences of most eukaryotic genes are interspersed with unexpressed regions. Because of this eukaryotic genes known as split genes.
Splicing reaction involves the removal of nonfunctional or non-coding introns and joining of functional or coding exons.
Exons:
They are the coding or functional sequences (or) expressed sequences of gene which gets transcribed in the primary RNA transcript and is retained in the final mature m-RNA.
Introns:
They are the noncoding or nonfunctional intervening sequences (IVs) of gene which gets transcribed in the primary RNA transcript but are not retained in the mature m-RNA as a result of splicing reactions.
Mechanism of splicing:
For splicing to occur the following sequences are necessary at the splice site junctions and in the introns. At the splice junction AAGU is the highly conserved sequence at the 5 boundary and AGG at the 3 boundary. In the introns, a conserved sequence of CURAY has been found about 20 to 50 residues upstream at the 3 splice site.
Steps:
i) First a 2-5 phosphodiester bond is formed between an introns adenosine residue and its Guanosines 5-terminal phosphate group with the concomitant release of the 5-exons. The introns thereby assume Lariat structure.
ii) The adenosine residue at the Lariat branch has been identified as the A in the CURAY sequence [where R represent purines and Y represent pyrimidines] which is highly conserved in vertebrate m-RNA.
iii) Now free 3-OH group of the 5-exon forms a phosphodiester bond with the 5-terminal phosphate of the 3-exon, yielding the splice product. The introns is eliminated in its Lariat form.
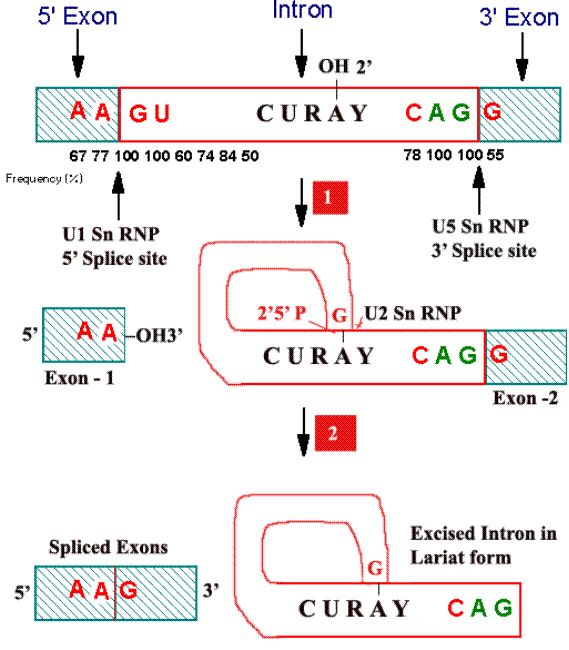
Role of SnRNPs:
Splicing reactions are mediated by small nuclear ribonucleoproteins [SnRNPs; pronounced as snurps] which are a complex of SnRNAs and proteins. There are more than 8 types of SnRNPs. Of which U1SnRNP, U2SnRNP, U5SnRNP, U4 and U6 SnRNPs are best characterized. U1SnRNP recognizes 5 splice junction, U2SnRNP then recognizes the introns region that forms the branch point while U5SnRNP recognizes the 3 splice junction.
SPLICEOSOME:
The large RNA-protein body, within which, nuclear m-RNA precursor is processed to remove introns. It is of 50S-60S particle. Spliceosome brings together a pre-m-RNA, the foregoing SnRNPs and a variety of pre-m-RNA binding proteins. Note that the spliceosome, which consists of 5 RNAs and atleast 50 polypeptides, is comparable in size and complexity to the E.Colis large ribosomal subunit 50S. Spliceosome carry out the splicing reactions of pre-m-RNA.
iv) Methylation:
During or shortly after the synthesis of vertebrate pre-m-RNAs, approximately 0.1% of their A residues are methylated at N6. These m6As tend to occur in the sequence RRm6ACX, where X is rarely G. Although the functional significance of these methylated As is unknown, it should be noted that a large fraction of them are components of the corresponding mature m-RNAs.
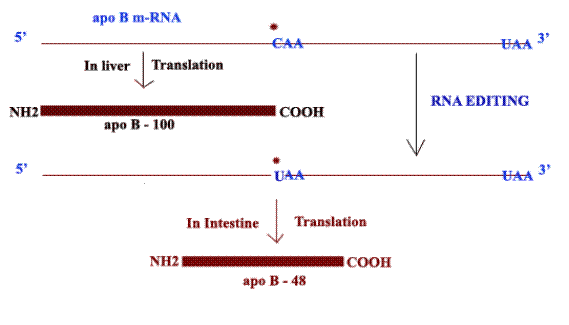
RNA EDITING:
RNA editing is a process in which the sequence of a pre-m-RNA altered. As a result, the sequence of the corresponding mature m-RNA differs from the exons encoding it in genomic DNA.
Certain m-RNAs from a variety of eukaryotic organisms have been found to differ from their corresponding genes in several unexpected ways, including CΰU and UΰC changes, the insertion or deletion of U residues and the insertion of multiple G or C residues.
RNA editing of apo-B m-RNA:
The apo-B m-RNA produced in the liver has the same sequences as the exons in the primary transcript. This m-RNA is translated into Apo B-100 whereas in the apo-B m-RNA produced in the intestine, the CAA codon in exon 26 is edited to a UAA stop codon. As a result, intestine cells produce Apo-B-48. Production of different apo-B from the same gene is because of RNA editing. Addition of U residues to m-RNA achieved with the help of guide RNAs (gRNA) during RNA editing.
Example: m-RNA of mitochondria of trypanosomes.
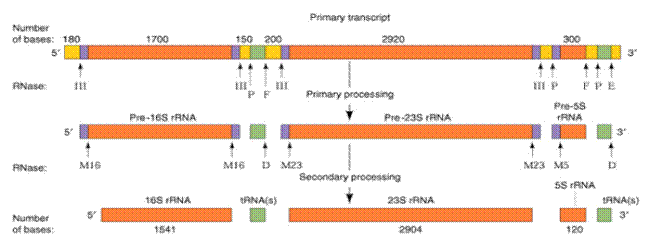
PROCESSING OF r-RNA:
Processing of prokaryotic r-RNA:
The prokaryotic r-RNAs are of three types namely
a) 16S r-RNA (1541 ribonucleotides)
b) 23S r-RNA (2904 ribonucleotides)
c) 5S r-RNA (120 ribonucleotides)
The primary transcript of these ribosomal RNAs is linked together as a nucleotide chain of more than 5,500 ribonucleotides. The primary r-RNA transcript undergoes processing in the following steps
i) Primary processing
ii) Secondary processing
iii) Methylation
i) Primary processing:
In this process, the primary rRNA undergoes cleavages in which the rRNAs (16S, 23S and 5S) and tRNAs are cleaved by trimming of the flanking nucleotide sequences. The trimming involves endonucleolytic cleavage which is catalyzed by RNase III, RNase P, RNase E and RNase F.
ii) Secondary processing:
The endonucleolytic activity of RNse III, P, E and F does not completely trim the flanking regions of the r-RNAs. The 5 and 3 ends of 16S r-RNA, 23S r-RNA and 5S r-RNA gets further trimmed by the RNase M16, M23 and M5 respectively and RNase D involves in the trimming of flanking regions of t-RNA. After secondary processing, the rRNAs get associated with proteins to form ribosomes.
iii) Methylation:
During ribosomal assembly, the 16S r-RNA and 23S r-RNA are methylated at a total of 24 specific nucleoside residues. The methylation reaction which employ S-adenosine methionine, a methyl donor, yield N6, N6 Dimethyl adenine and 2-o-methyl ribose residues which are thought to protect adjacent phosphodiester bond from degradation by intracellular RNases. This is because RNases hydrolysis involves utilization of the free 2-OH groups of ribose. However, the function of base methylation is unknown.
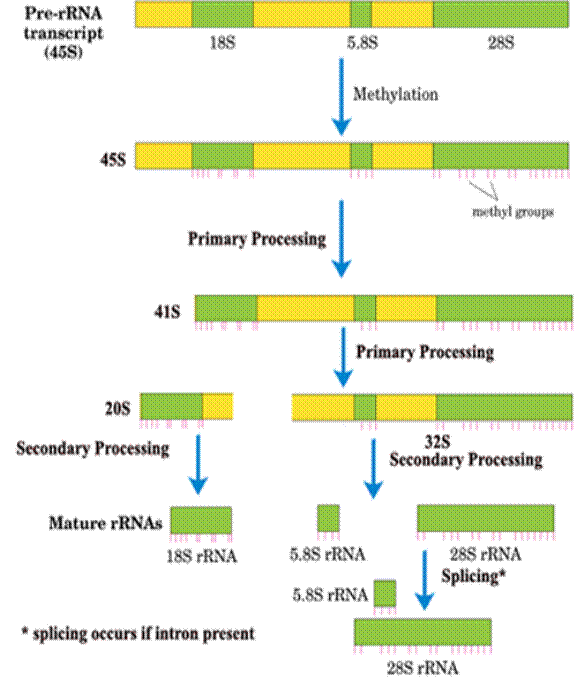
Processing of Eukaryotic r-RNA:
The eukaryotic r-RNAs are of four types namely
a) 18S r-RNA (1900 nucleotides)
b) 5.8S r-RNA (160 nucleotides)
c) 28S r-RNA (4700 nucleotides)
d) 5S r-RNA (120 nucleotides)
The Primary r-RNA transcript is of approximately 13,000 nucleotide residues and has a sedimentation coefficient of 45S. Starting from 5-end, the structural arrangement of various r-RNAs in the pre-r-RNA is as follows:
5 --- 18S r-RNA ---- 5.8S r-RNA ----28S r-RNA ----3
As in prokaryotes, these r-RNAs separated by spacer sequences. Processing of these r-RNAs involved in the following steps
i) Methylation
ii) Primary processing
iii) Secondary processing
iv) Splicing
i) Methylation:
In the first stage of its processing, 45S r-RNA is specifically methylated at approximately 110 sites that occur mostly in its r-RNA sequences. About 80% of these modifications yield O2 methyl ribose residues and the remainder form methylated bases such as N6, N6 Dimethyladenine and 2-methylguanine. After, methylation, pre r-RNA undergoes other stages of processing.
ii) Primary processing:
After methylation, 45S r-RNA undergoes cleavage of 5-end spacer to yield 41S r-RNA. The next step involves cleaving 41S r-RNA into two pieces, 32S and 20S that contains the 28S and 18S sequences respectively. The 32S precursor also retains the 5.8S RNA sequence. This ends primary processing stage.
iii) Secondary processing:
In this stage, 32S precursor is split to yield the mature 28S and 5.8S RNAs, which base pair with each other and the 20S precursor is trimmed to mature 18S size.
iv) Splicing:
Only a few eukaryotic r-RNA genes contain introns. So, they alone undergo splicing.
Example:
26S part of the protozoan Tetrahymena thermophilia r-RNA precursor does contain introns and it can be spliced by 26S r-RNA itself without any help from proteins. The Tetrahymena 26S r-RNA is the equivalent of the mammalian 28S r-RNA.
Mechanism of splicing:
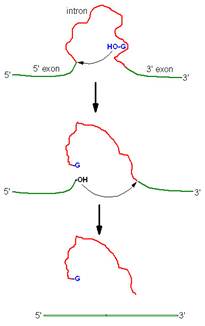
Group I introns splicing:
Splicing of 26S r-RNA examined and explained by Thomas Cech. The introns in 26S r-RNA of Tetrahymena are known as group I introns which also occur in the nuclei, mitochondria and chloroplast of diverse eukaryotes (although not vertebrates). In the first step of splicing, a guanine nucleotide attacks the adenine nucleotide residue at the 5-end of the introns, releasing exon-1 from the rest of the molecule and leaving introns -1 and exon -2 complexes.
In the second step, exon-1 attacks exon-2, performing the splice reaction that releases linear introns and joins the two exons together.
Group II introns splicing:
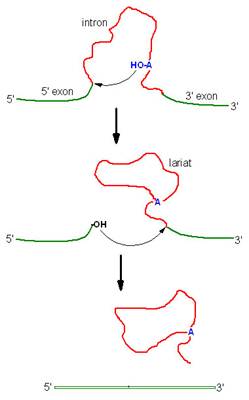
Introns of yeast mitochondrial pre-r-RNA are known as group-II introns which also occur in the mitochondria of fungi and plants and comprise the majority of the introns in chloroplasts. Group II introns also self splice but they do not need assistance from guanosine to start the reaction. Instead, the initiating entity is an adenosine nucleotide residue within the introns of the RNA itself.
In the first step, 2-OH of adenosine residue attacks the 5-end nucleotide residue to form lariat structure with introns-1 and exon-2 complex and release exon-1.
In the second step, exon-1 attacks exon-2, performing splice reaction tat release lariat introns and joins the two exons together.
The details of this r-RNA processing scheme are not universal. Even the mouse does things a little differently and the frog precursor is only 40S which is quite a bit smaller than 45S. Still, the basic mechanism of r-RNA processing, including the order of mature sequences in the precursor is preserved throughout the eukaryotic kingdom.
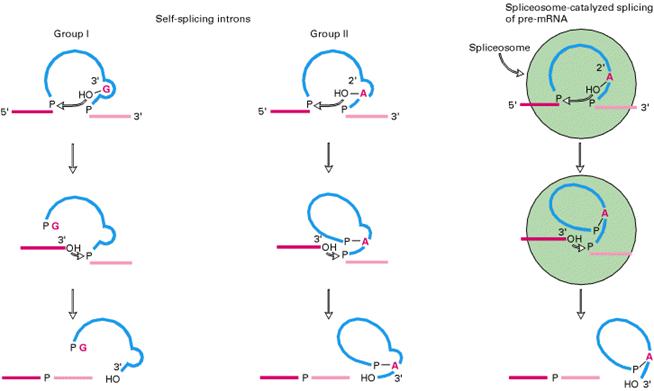
Ribozymes:
RNAs with enzymatic activities are referred as ribozymes.
Example: Hammerhead ribozymes of plant virus and Tetrahymena thermophilia r-RNA.
Since splicing carried out by RNA itself, the process is known as self splicing.
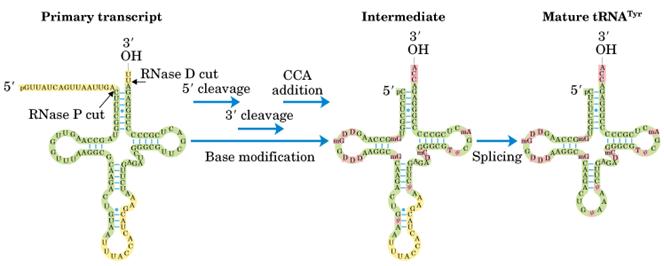
Processing of t-RNA:
Both prokaryotic and eukaryotic pre t-RNA undergo post transcriptional modification. The steps for post transcriptional modification of pre-t-RNA are as follows:
i) First, the flanking regions of the 3-OH and 5 phosphate ends are cleaved by the endonuclease action of RNase D and RNase P respectively.
ii) Then, the introns in the anticodon loop space, spliced out by splicing reaction.
iii) Tri nucleotide CCA is added to the 3-end to give 32-OH ACC terminus. This reaction is catalyzed by t-RNA specific nucleotidyl transferase. It is unique reaction because this enzyme catalyzes transfer of 3 nucleoside phosphodiester bond formation in one step.
iv) Finally, the t-RNA undergoes base modifications to give mature t-RNA.
Splicing mechanism:
Splicing mechanism in pre-t-RNA differ from mechanisms utilized by self splicing introns and spliceosomes.
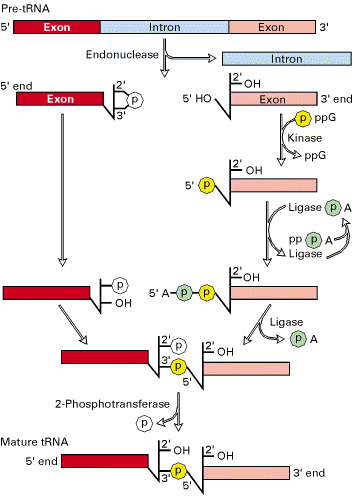
Splicing reactions requires four enzymes namely t-RNA specific endonuclease, cyclic phosphodiesterase, t-RNA specific ligase and 2-phosphotransferase.
By the action of endonuclease, introns removed. Following, excision of introns, a 2-3 cyclic phosphomonoester bond forms on the cleaved end of the 5-exon. The multi step reaction joining the two exon requires two nucleoside triphosphates: a GTP, which contributes the phosphate group for the 3ΰ5 linkage in the finished t-RNA molecule and an ATP, which forms an activated ligaseAMP intermediate. The 2 phosphate on the 5-exon is removed in the final step.
INHIBITORS OF RNA METABOLISM:
A large variety of inhibitors of RNA synthesis have been identified. The inhibitors fall into three groups. They being
i) Inhibitors act by binding to DNA
ii) Inhibitors act by binding to RNA polymerase
iii) Inhibitors act by binding to RNA chain
i) Inhibitors act by binding to DNA:
The best known example of inhibitors that bind to DNA is Actinomycin D, an antibiotic produced by streptomyces antiboticus. The inhibition of RNA synthesis is caused by the insertion (interaction) of its phenoxazone ring between two G-C pairs, with the side chains projecting into the minor groove of the double helix, hydrogen bonded to guanosine residues. RNA Polymerase binding to DNA that contains Actinomycin D is only slightly impaired, but RNA chain elongation in both eukaryotes and prokaryotes is blocked.
Ethidium bromide also intercalates into DNA and at low concentrations preferentially binds to negatively supercoiled DNA. It has been used to selectively inhibit transcription in mitochondria which contains supercoiled DNA.
ii) Inhibitors act by binding to RNA Polymerase:
Rifampicin is a synthetic derivative of a naturally occurring antibiotic, Rifampicin that inhibits bacterial DNA dependent RNA polymerase but not T7 RNA polymerase or eukaryotic RNA polymerase. It binds tightly to the beta subunit. Although it does not prevent promoter binding or formation of the first phosphodiester bond, it effectively prevents synthesis of longer RNA chains. It does not inhibit elongation when added after initiation has occurred.
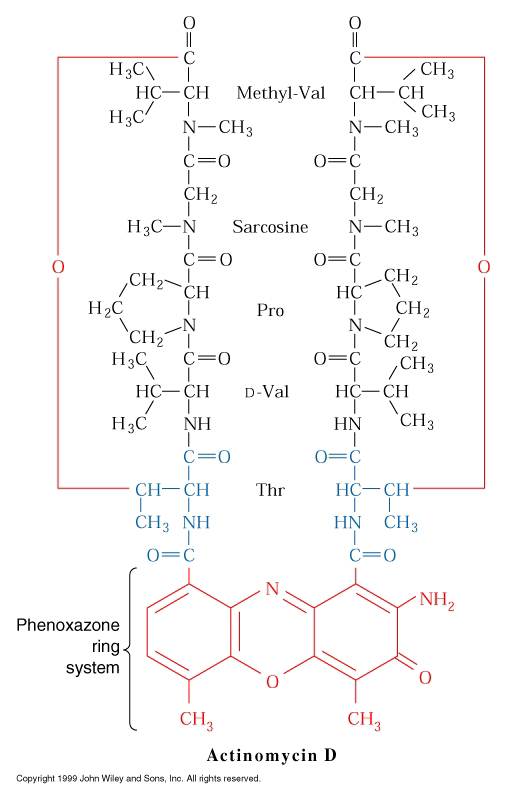
Another antibiotic, streptolydigin, also binds to the beta subunit, it inhibits all bond formation.
The most useful inhibitors of eukaryotic transcription have been a-amanitin, a major toxic substance in the poisonous mushroom Amanita phalloides. The toxin preferentially binds to and inhibits RNA Pol-II. At high concentrations it also can inhibit RNA Pol-III but not RNA Pol-III but not RNA Pol-I or bacterial, mitochondrial or chloroplast RNA Polymerases.
iii) Inhibitors act by binding to growing RNA chain:
Cordycepin in its 5-triphosphorylated form is a substrate analog that is incorporated into growing RNA chains by most RNA polymerases. It causes chain termination after incorporation, since it does not contain the 3-hydroxyl group necessary for the formation of the next phosphodiester bond.
Inhibition like Nalidixic acid, Novobiocin and Dichloro ribo benzene (DRB) etc., also inhibit transcription.
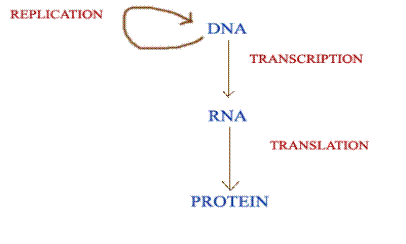
CENTRAL DOGMA OF MOLECULAR BIOLOGY:
Central dogma of Molecular Biology states that the genetic information can flow from DNA to DNA, DNA to RNA and RNA to protein only. It is represented as below:
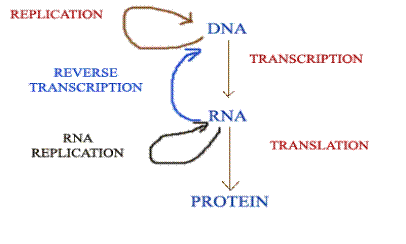
MODIFIED CENTRAL DOGMA:
The discovery of reverse transcriptase has modified the central dogma of molecular biology which held that genetic information should pass only from DNA to RNA. This enzyme, synthesis DNA forms RNA, thus showing that the information can flow from RNA to DNA.
Like reverse transcriptase, RNA replicase also has modified the central dogma molecular biology. Normally, information transferred from DNA to RNA by Transcription and RNA to DNA by reverse transcriptase but RNA replicase transfer the information from RNA to RNA. Thus it modifies the dogma of molecular biology. Modification by RNA replicase is as follows:
Methylation of DNA
The importance of methylation in DNA-protein interactions is well known. A small percentage of cytosine residues are methylated in many eukaryotic organisms, mainly in CpG sequences (see fig. 13.3); 80% of the cytosines in CpG sequences in human DNA are methylated. (Often, when we refer to a sequence of two bases on the same strand of DNA, we put a p between themCpGto indicate that they are on the same strand connected by a phosphodiester bond and not on two different strands as a hydrogen-bonded base pair.) The degree of methylation of DNA is related to the silencing of a gene. Genes that are dormant in one cell type but active in another, or genes that are dormant at one stage of development but active in another, are usually less methylated when active and more fully methylated when inactive. For example, adenovirus, a cancer-causing virus, has been observed in many eukaryotic cell lines. In most lines in which the adenovirus DNA has integrated into the host chromosome, late viral genes are turned off. These genes are highly methylated at their CCGG or GCGC sites. In addition, chemicals that prevent methylation frequently activate previously dormant genes. For example, 5-azacytidine inhibits methylation; X chromosomal genes, which are normally deactivated, can be reactivated by treatment with 5-azacytidine. There are numerous other examples of the activation of genes after treatment with this chemical. The activated genes lack methylated cytosines that were previously methylated. Finally, the possibility exists that DNA methylation can affect the pattern of chromatin structure.
Recent work has also indicated that the methylation itself may not prevent transcription, but rather may be a signal for transcriptional inactivity. In the thale cress plant,Arabidopsis thaliana, a protein named Mom (for Morpheus molecule), has been discovered that, when mutated, results in genes that have heavy methylation levels but are actively transcribed. Thus, the methylation level can be separated from the transcriptional activity of genes, although the two usually occur together. Arabidopsis is proving to be a good model in the study of the role of methylation in transcriptional activation because other common model organisms, namely fruit flies, yeast, and the nematode, Caenorhabditis elegans, do not have methylation of their DNA.
Further interest has been generated in the role of methylation in controlling gene expression by the discovery of Z DNA, and the fact that Z DNA can be stabilized by methylation (see chapter 9). This observation has led to a model of transcriptional regulation based on alternative DNA structures. Sequences (such as CpG repetitions) that could exist as Z DNA exist as B DNA when being transcribed. If the gene is to be silenced (turned off), the CpG sequences are converted to stable Z DNA by methylation, which then blocks transcription.This possibility has gained some interest because of the recent discovery of an enzyme, double-stranded RNA adenosine deaminase (ADAR1), that binds to Z DNA sequences.
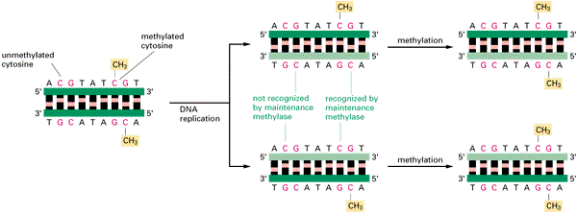
How DNA methylation patterns are
faithfully inherited. In vertebrate DNAs a large
fraction of the cytosine nucleotides in the sequence CG are methylated (see
Figure 9-67). Because of the existence of a methyl-directed methylating
enzyme (the maintenance methylase), once a pattern of DNA methylation is
established, each site of methylation is inherited in the progeny DNA, as
shown. This means that changes in DNA methylation patterns will be perpetuated
in all of the progeny of a cell.
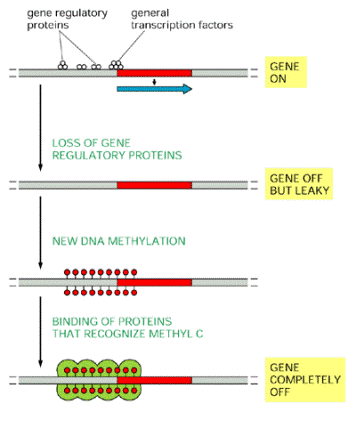
How DNA methylation may help turn off
genes. The binding of gene regulatory proteins and
general transcription factors near an active promoter prevents DNA methylation
by some unknown mechanism. If most of these sequence-specific DNA-binding
proteins dissociate, however, as generally occurs when a gene is turned off,
the DNA becomes methylated, which enables other proteins to bind, and these
shut down the gene completely.
GENE EXPRESSION
The controls that act on gene expression (i.e., the ability of a gene to produce a biologically active protein) are much more complex in eukaryotes than in prokaryotes. A major difference is the presence in eukaryotes of a nuclear membrane, which prevents the simultaneous transcription and translation that occurs in prokaryotes. Whereas, in prokaryotes, control of transcriptional initiation is the major point of regulation, in eukaryotes the regulation of gene expression is controlled nearly equivalently from many different points.
GENE CONTROL
IN PROKARYOTES
In bacteria, genes are clustered into operons (gene clusters) that encode the proteins necessary to perform coordinated function, such as biosynthesis of a given amino acid. RNA that is transcribed from prokaryotic operons is polycistronic, a term implying that multiple proteins are encoded in a single transcript.
In bacteria, control of the rate of transcriptional initiation is the predominant site for control of gene expression. As with the majority of prokaryotic genes, initiation is controlled by two DNA sequence elements that are approximately 35 bases and 10 bases, respectively, upstream of the site of transcriptional initiation and as such are identified as the -35 and -10 positions. These 2 sequence elements are termed promoter sequences, because they promote recognition of transcriptional start sites by RNA polymerase. The consensus sequence for the -35 position is TTGACA, and for the -10 position, TATAAT. (The -10 position is also known as the Pribnow-box.) These promoter sequences are recognized and contacted by RNA polymerase.
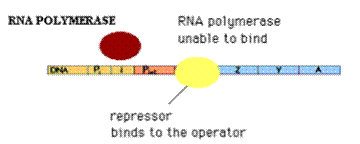
The activity of RNA polymerase at a given promoter is in turn regulated by interaction with accessory proteins, which affect its ability to recognize start sites. These regulatory proteins can act both positively (activators) and negatively (repressors). The accessibility of promoter regions of prokaryotic DNA is in many cases regulated by the interaction of proteins with sequences termed operators. The operator region is adjacent to the promoter elements in most operons and in most cases the sequences of the operator bind a repressor protein. However, there are several operons in E. coli that contain overlapping sequence elements, one that binds a repressor and one that binds an activator.
Regulatory sequences like the operator are called cis-acting control elements, because they affect the expression of only linked genes on the same DNA molecule. On the other hand, proteins like the repressor are called trans-acting factors because they can affect the expression of genes located on other chromosomes within the cell.
Prokaryotic genes that encode the proteins necessary to perform coordinated function are clustered into operons. Two major modes of transcriptional regulation function in bacteria (E. coli) to control the expression of operons. Both mechanisms involve repressor proteins. One mode of regulation is exerted upon operons that produce gene products necessary for the utilization of energy; these are catabolite-regulated operons. The other mode regulates operons that produce gene products necessary for the synthesis of small biomolecules such as amino acids. Expression from the latter class of operons is attenuated by sequences within the transcribed RNA. Trp operon is an example for such operon.
OPERON
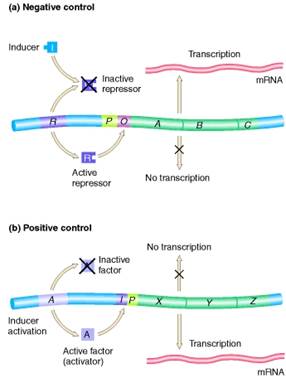
The operon model of prokaryotic gene regulation was proposed by Fancois Jacob and Jacques Monod. Groups of genes coding for related proteins are arranged in units known as operons. An operon consists of an operator, promoter, regulator, and structural genes. The regulator gene codes for a repressor protein that binds to the operator, obstructing the promoter (thus, transcription) of the structural genes. The regulator does not have to be adjacent to other genes in the operon. If the repressor protein is removed, transcription may occur. Operons are either inducible or repressible according to the control mechanism. Seventy-five different operons controlling 250 structural genes have been identified for E. coli. Both repression and induction are examples of negative control since the repressor proteins turn off transcription.
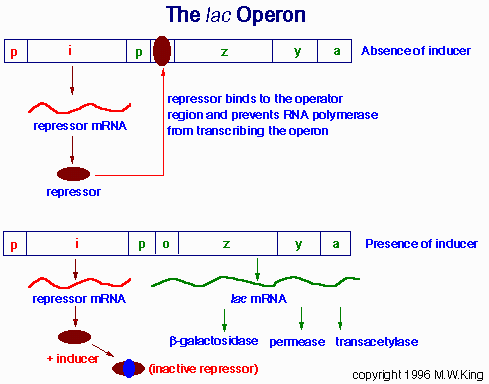
LAC OPERON (INDUCIBLE SYSTEM)
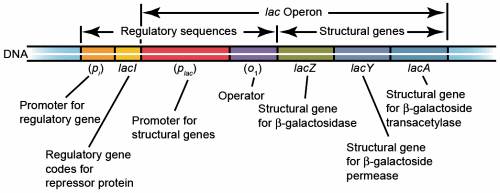
The lac operon consists of three structural genes, and a promoter, a terminator, regulator, and an operator. The three structural genes are: lacZ, lacY, and lacA.
· lacZ encodes β-galactosidase (LacZ), an intracellular enzyme that cleaves the disaccharide lactose into glucose and galactose.
· lacY encodes β-galactoside permease (LacY), a membrane-bound transport protein that pumps lactose into the cell.
· lacA encodes β-galactoside transacetylase (LacA), an enzyme that transfers an acetyl group from acetyl-CoA to β-galactosides.
Only lacZ and lacY appear to be necessary for lactose catabolism.
Specific control of the lac genes depends on the availability of the substrate lactose to the bacterium. The proteins are not produced by the bacterium when lactose is unavailable as a carbon source. The lac genes are organized into an operon; that is, they are oriented in the same direction immediately adjacent on the chromosome and are co-transcribed into a single polycistronic mRNA molecule. Transcription of all genes starts with the binding of the enzyme RNA polymerase (RNAP), a DNA-binding protein, which binds to a specific DNA binding site, the promoter, immediately upstream of the genes. From this position RNAP proceeds to transcribe all three genes (lacZYA) into mRNA.
The first control mechanism is the regulatory response to lactose, which uses an intracellular regulatory protein called the lactose repressor to hinder production of β-galactosidase in the absence of lactose. The lacI gene coding for the repressor lies nearby the lac operon and is always expressed (constitutive). If lactose is missing from the growth medium, the repressor binds very tightly to a short DNA sequence just downstream of the promoter near the beginning of lacZ called the lac operator. The repressor binding to the operator interferes with binding of RNAP to the promoter, and therefore mRNA encoding LacZ and LacY is only made at very low levels. When cells are grown in the presence of lactose, however, a lactose metabolite called allolactose, which is a combination of glucose and galactose, binds to the repressor, causing a change in its shape. Thus altered, the repressor is unable to bind to the operator, allowing RNAP to transcribe the lac genes and thereby leading to high levels of the encoded proteins. Isopropyl β-D-1-thiogalactopyranoside, abbreviated IPTG, is a molecular biology reagent. This compound is used as a molecular mimic of allolactose, a lactose metabolite that triggers transcription of the lac operon. Unlike allolactose, the sulfur (S) atom creates a chemical bond which is non-hydrolyzable by the cell, preventing the cell from "eating up" or degrading the inductant; therefore the IPTG concentration remains constant. For induction, a sterile 1 M solution of IPTG is typically added by 1:1000 dilutions into a logarithmically growing bacterial culture. Different final concentration of IPTG may be used.
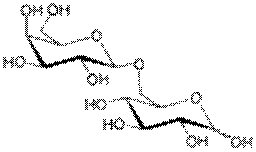
Allolactose
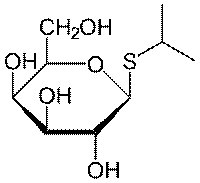
Isopropyl β-D-1-thiogalactopyranoside
The second control mechanism is a response to glucose, which uses the catabolite activator protein (CAP also known as the cAMP receptor protein (CRP)) to greatly increase production of β-galactosidase in the absence of glucose. Cyclic adenosine monophosphate (cAMP) is a signal molecule whose prevalence is inversely proportional to that of glucose. It binds to the CAP, which in turn allows the CAP to bind to the CAP binding site (a 16 bp DNA sequence upstream of the promoter on the left), which assists the RNAP in binding to the DNA. In the absence of glucose, the cAMP concentration is high and binding of CAP-cAMP to the DNA significantly increases the production of β-galactosidase, enabling the cell to hydrolyse (digest) lactose and release galactose and glucose.
cAMP-CRP binds to the lac operon just upstream of the promoter. In this position, a molecule of cAMP-CRP can assist RNA polymerase to bind by direct protein-protein contacts:
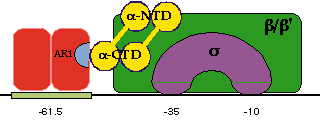
In this "cartoon" picture, a CRP dimer is positioned at its binding site, which is centred 61.5 bp upstream of the startpoint of transcription. One part of the dimer (labelled AR1) makes a direct contact with the carboxy-terminal domain of the alpha subunit of RNA polymerase thus helping it bind at the promoter.
Positive control
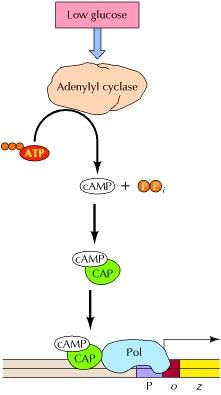
Positive control is the regulation mediated by a protein that is required for the activation of a transcription unit. Glucose repression (generally called catabolite repression) is now known to be mediated by a positive control system, which is coupled to levels of cyclic AMP (cAMP). In bacteria, the enzyme adenylyl cyclase, which converts ATP to cAMP, is regulated such that levels of cAMP increase when glucose levels drop. cAMP then binds to a transcriptional regulatory protein called catabolite activator protein (CAP). The binding of cAMP stimulates the binding of CAP to its target DNA sequences, which in the lac operon are located approximately 60 bases upstream of the transcription start site. CAP then interacts with the α subunit of RNA polymerase, facilitating the binding of polymerase to the promoter and activating transcription.
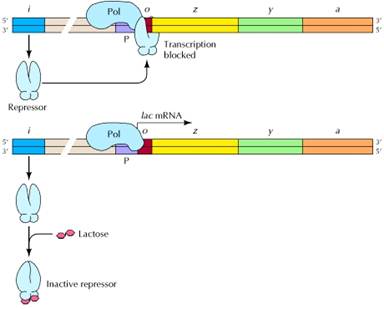
Negative control
Negative control is the regulation mediated by factors that block or turn off transcription. The i gene encodes a repressor which, in the absence of lactose, binds to the operator (o) and blocks transcription of the three structural genes (z, β-galactosidase; y, permease; and a, transacetylase). Lactose induces expression of the operon by binding to the repressor (bottom), which prevents the repressor from binding to the operator.
Negative control of the lac operon by glucose is achieved by a somewhat indirect mechanism. As glucose levels fall, intracellular levels of cyclic adenosine monophosphate (cAMP) begin to rise. When this happens, an intracellular protein called Catabolite Activator Protein (CAP) binds to the cAMP and becomes a positive-acting regulator of the lac operon, binding upstream from the -35 promoter sequence. This process was named catabolite repression before the detailed mechanism was well understood. Products of the metabolism of glucose (catabolites) suppress cAMP levels, which in turn prevents binding of the CAP-cAMP complex and thus "represses" transcription of the operon through failure to stimulate that transcription.
DUAL REGULATORY SYSTEMS
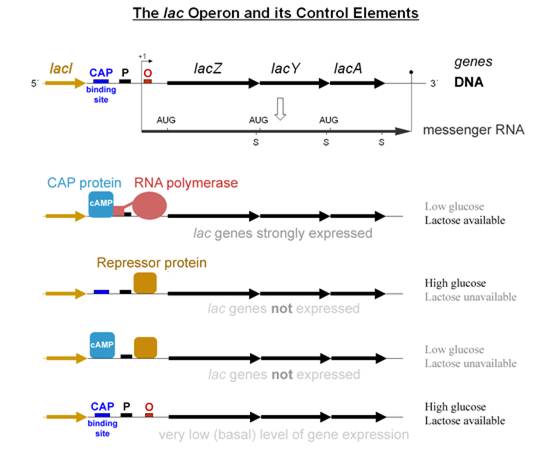
The lactose operon is subject both to negative control and to positive control. The lac repressor negatively regulates expression; cAMP-CRP positively activates expression. There are, as a result, four basic states of expression of the lac operon:
No Glucose and No Lactose
Under these conditions, there will be a high [cAMP] in the cell and CRP will be bound at its binding site upstream of the lac promoter. It will assist RNA polymerase to bind to the promoter but it will not activate transcription because the lactose repressor will remain bound to the operator sites since there is no inducer present. There will be essentially no transcription of the lac operon.
Without sugar substrates the cell cannot carry out much metabolism; however, it remains poised to use whatever it can whenever it can. In this case, if lactose does become available, the cell can and will immediately respond:
o Lactose permease will transport the lactose into the cell.
o RNA polymerase is positioned to start the expression of b-galactosidase so that the lactose can be utilized immediately.
Glucose Present but No Lactose
Under these conditions, there will be a low [cAMP] in the cell so CRP will not be bound at the lac promoter. Glucose transport also leads to direct inhibition of the lactose permease. In addition, the activity of lactose permease will be inhibited. There will be no transcription of the lac operon. As long as glucose is present in the growth medium there is little need to metabolize lactose and since lactose is not present there is no need to transport lactose into the cell or to express the genes of the lac operon.
Glucose and Lactose Present
Under these conditions, there will be a low [cAMP] in the cell so CRP will not be bound at the lac promoter. Glucose transport also leads to direct inhibition of the lactose permease. Lactose permease will be inhibited but some lactose will still enter the cell. There will be a low level transcription of the lac operon. As long as glucose is present in the growth medium there is little need to metabolize lactose. However, since lactose is now present, the cell would be foolish to ignore a sugar supply completely. The lac operon will be induced but, since CRP is not bound, the amount of transcription is relatively low.
Lactose Present but No Glucose
Under these conditions, there will be a high [cAMP] in the cell so CRP will be bound at the lac promoter. Lactose permease is not inhibited, so it will transport the lactose into the cell. There will be maximal transcription of the lac operon. With lactose as the sole sugar source, the cell must use every available molecule for its own benefit. Thus the lactose permease transport system will transport lactose into the cell and the lac operon will be both induced and activated.
The presence of two separate control systems allows the cell to respond more sensitively to the needs imposed by changing growth conditions. Many bacterial operons have dual control systems.
TRP OPERON
(REPRESSIBLE SYSTEM)
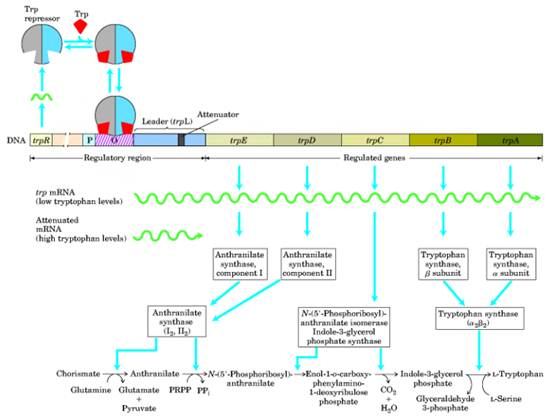
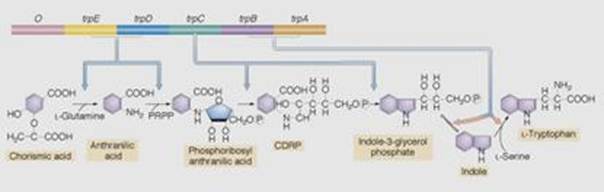
The inducible operons are activated when the substrate that is to be catabolized enters the cell. Anabolic operons function in the reverse manner: They are turned off (repressed) when their end product accumulates beyond the needs of the cell.Two entirely different, although not mutually exclusive,mechanisms seem to control the transcription of repressible operons.The first mechanism follows the basic scheme of inducible operons and involves the end product of the pathway. The second mechanism involves secondary structure in messenger RNA transcribed from an attenuator region of the operon. One of the best-studied repressible systems is the tryptophan, or trp, operon in E. coli. The trp operon contains the five genes that code for the synthesis of the enzymes that build tryptophan, starting with chorismic acid. It has a promoter-operator sequence ( p,o ) as well as its own regulator gene (trpR).
OPERATOR CONTROL
In this repressible
system, the product of the trpR gene,
the repressor, is inactive by itself; it does not recognize the operator
sequence of the trp operon. The
repressor only becomes active when it combines with tryptophan. Thus, when
tryptophan builds up, enough is available to bind with and activate the
repressor. Tryptophan is thus referred to as the corepressor. The
corepressor-repressor complex then recognizes the operator, binds to it, and
prevents transcription by RNA polymerase. After the available tryptophan in the
cell is used up, the diffusion process causes tryptophan to leave the
repressor, which then detaches from the trp
operator. The transcription process no longer is blocked and can proceed
normally (the operon is now derepressed). Transcription continues until
enough of the various enzymes have been synthesized to again produce an excess
of tryptophan. Some becomes available to bind to the repressor and make a
functional complex, and the operon is again shut off and the process repeated,
ensuring that
tryptophan is being
synthesized as needed (fig.).
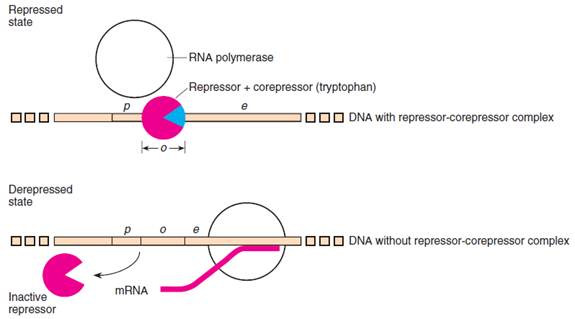
Figure: The repressor-corepressor complex binds at the operator and prevents the transcription of the trp operon in E. coli. Without the corepressor, the repressor cannot bind, and therefore transcription is not prevented. The blue wedge is the corepressor (two tryptophan molecules), and the partial red circle is the repressor.
ATTENUATOR
CONTROL
Details of the second control mechanism of repressible operons have been elucidated primarily by C. Yanofsky and his colleagues, who worked with the tryptophan operon in E. coli. This type of operon control, control by an attenuator region, has been demonstrated for at least five other amino acid-synthesizing operons, including the leucine and histidine operons. This regulatory mechanism may be the same for most operons involved in the synthesis of an amino acid. In the trp operon, an attenuator region lies between the operator and the first structural gene. The messenger RNA transcribed from the attenuator region, termed the leader transcript, has been sequenced, revealing two surprising and interesting facts. First, four subregions of the messenger RNA have base sequences that are complementary to each other so that three different stem-loop structures can form in the messenger RNA (fig. 14.14). Depending on circumstances, regions 12 and 34 can form two stem-loop structures, or region 23 can form a single stem-loop. When one stemloop structure is formed, the others are preempted.
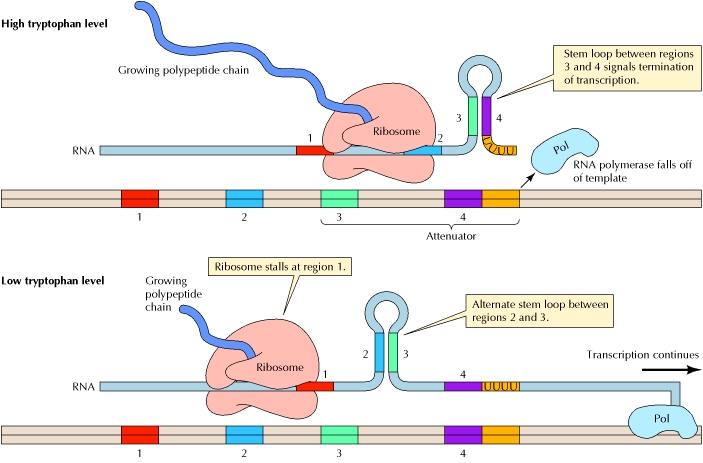
The trp
mRNA is translated while still being synthesized. The mechanism of attenuation
depends on the fact that translation in bacteria is coupled with transcription,
so ribosomes begin translating the 5′ end of an mRNA while it is still
being synthesized. Thus, the rate of translation can affect the structure of
the growing RNA chain, which in turn determines whether further transcription
can continue. Transcription termination is signaled by a stem-loop structure
that forms by complementary base pairing between two specific sequences of the
growing Trp mRNA
chain. In the presence of high levels of tryptophan, the ribosomes proceed
along the message slightly behind the site of transcription. Under these
conditions, the mRNA regions designated 3 and 4 hybridize to form a stem-loop
structure that signals the termination of transcription. In the presence of low
levels of tryptophan, however, the ribosomes stall at region 1 of the mRNA,
which contains two adjacent codons for tryptophan. In this case, since region 2
is not bound to a ribosome, it is free to form an alternative stem-loop
structure by hybridizing to region 3. This hybridization prevents formation of
the 34 stem loop, and transcription is able to continue past the attenuator
sequence. The stem-loop 23 structure is
referred to as the preemptor stem. Note
that the preemptor stem is not a rho-independent transcription terminator and
thus, without the rho protein present, will not terminate transcription. The
critical region of Trp
mRNA contains two adjacent tryptophan codons, so the rate of translation is
highly dependent on tryptophan levels; this is the link between transcriptional
attenuation and the availability of tryptophan. If tryptophan levels in the
cell are low, the ribosome stalls at this point and transcription of Trp mRNA continues. If
tryptophan is abundant, translation continues and transcription is terminated.
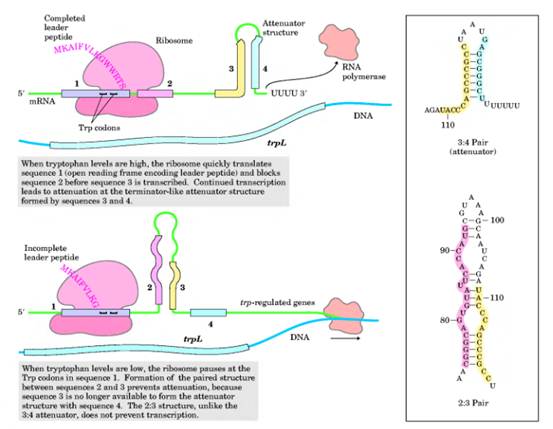
Excess
Tryptophan
Assuming that the operator site is available to RNA polymerase, transcription of the attenuator region will begin. As soon as the 5_ end of the messenger RNA for the leader peptide gene has been transcribed, a ribosome attaches and begins translating this messenger RNA. Depending on the levels of amino acids in the cell, three different outcomes can take place. If the concentration of tryptophan in the cell is such that abundant tryptophanyl-tRNAs exist, translation proceeds down the leader peptide gene. The moving ribosome overlaps regions 1 and 2 of the transcript and allows stem-loop 34 to form, as shown in the configuration at the far left of figure 14.16. This stem-loop structure, referred to as the terminator, or attenuator, stem, causes transcription to be terminated. Note that stem-loop 34, the terminator stem, followed by a series of uracil-containing bases, is a rho-independent transcription terminator. Hence, when existing quantities of tryptophan, in the form of tryptophanyl-tRNA, are adequate for translation of the leader peptide gene, transcription is terminated.
Tryptophan
Starvation
If the quantity of tryptophanyl-tRNA is lowered, the ribosome must wait at the first tryptophan codon until it acquires a Trp-tRNATrp. This is shown in the configuration in the middle part of figure 14.16. The stalled ribosome will permit stem-loop 23 to form, which precludes the formation of the terminator stem-loop (34). In this configuration, transcription is not terminated, so that eventually, the whole operon is transcribed and translated, raising the level of tryptophan in the cell.The stem-loop 23 structure is referred to as the preemptor stem. Note that the preemptor stem is not a rho-independent transcription terminator and thus, without the rho protein present, will not terminate transcription.
TRAP
Control
The tryptophan operon in bacilli such as Bacillus subtilis is also controlled by attenuation, but secondary structure in the mRNA transcript is induced by binding not the ribosome, but a trp RNA-binding attenuation protein (TRAP). This protein attaches to the nascent messenger RNA only after the protein binds tryptophan molecules; the result is a terminator stem that forms in the messenger RNA. In the absence of excess tryptophan,TRAP does not bind to the messenger RNA, a preemptor (also called an antiterminator) stem, not the terminator stem, forms, and transcription continues. Recently, the structure of the protein was worked out; it has eleven symmetrical loops, each of which can bind a tryptophan molecule.When TRAP is bound to tryptophan molecules, it can attach to triplets in the messenger RNA transcript, triplets of GAG or UAG. The TRAP wraps the mRNA around itself, forming an elegant pinwheel.
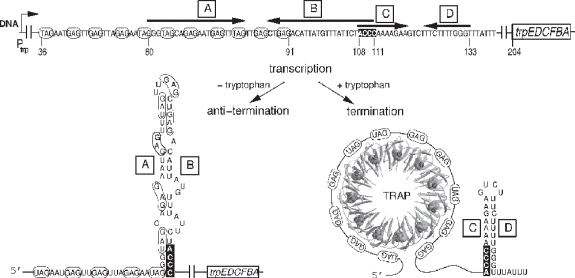
Redundant
Controls
Some amino acid
operons are controlled only by attenuation, such as the his
operon in E. coli, in which the leader peptide gene contains seven
histidine codons in a row, or the trp operon
in B. subtilis. Redundant control (repression and attenuation) of
tryptophan biosynthesis in E. coli allows the cell to test both the
tryptophan levels (tryptophan is the corepressor) and the tryptophanyltRNA
levels (in the attenuator control system).The
attenuator system also allows the cell to regulate tryptophan synthesis on the
basis of the shortage of other amino acids. For example, when there is a
shortage of tryptophan and arginine, operator control allows transcription to
begin, but attenuator control terminates transcription because stem-loops 12
and 34 form.
Alternate Splicing
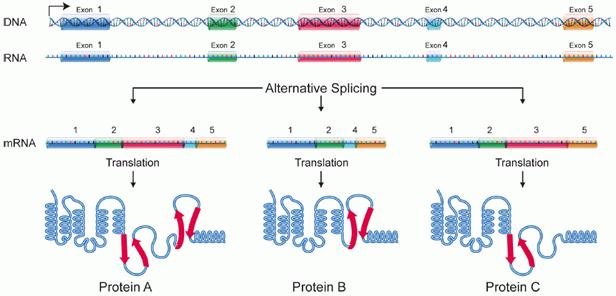
Alternative splicing is a regulated
process during gene expression that results in a single gene coding for
multiple proteins. In this process, particular exons of a gene may be included
within or excluded from the final, processed messenger RNA (mRNA) produced from
that gene. Consequently, the proteins translated from alternatively spliced
mRNAs will contain differences in their amino acid sequence and, often, in
their biological functions (see Figure). Notably, alternative splicing allows
the human genome to direct the synthesis of many more proteins than would be
expected from its 20,000 protein-coding genes. Alternative splicing is
sometimes termed differential splicing.
Modes of splicing
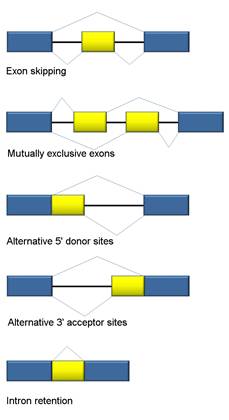
Five basic modes of alternative splicing are generally recognized.
- Exon skipping or cassette exon: in this case, an exon may be spliced out of the primary transcript or retained. This is the most common mode in mammalian pre-mRNAs.
- Mutually exclusive exons: One of two exons is retained in mRNAs after splicing, but not both.
- Alternative donor site: An alternative 5' splice junction (donor site) is used, changing the 3' boundary of the upstream exon.
- Alternative acceptor site: An alternative 3' splice junction (acceptor site) is used, changing the 5' boundary of the downstream exon.
- Intron retention: A sequence may be spliced out as an intron or simply retained. This is distinguished from exon skipping because the retained sequence is not flanked by introns. If the retained intron is in the coding region, the intron must encode amino acids in frame with the neighboring exons, or a stop codon or a shift in the reading frame will cause the protein to be non-functional. This is the rarest mode in mammals.
In addition to these primary modes of alternative splicing, there are two other main mechanisms by which different mRNAs may be generated from the same gene; multiple promoters and multiple polyadenylation sites. Use of multiple promoters is properly described as a transcriptional regulation mechanism rather than alternative splicing; by starting transcription at different points, transcripts with different 5'-most exons can be generated. At the other end, multiple polyadenylation sites provide different 3' end points for the transcript. Both of these mechanisms are found in combination with alternative splicing and provide additional variety in mRNAs derived from a gene.
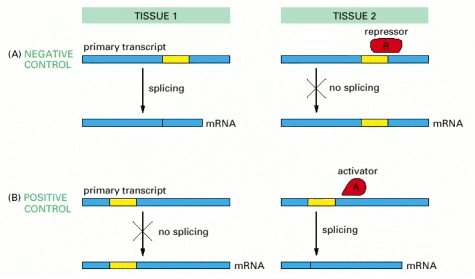
Regulation of splicing
RNA splicing can be regulated either negatively, by a regulatory molecule that prevents the splicing machinery from gaining access to a particular splice site on the RNA, or positively, by a regulatory molecule that helps direct the splicing machinery to an otherwise overlooked splice site. In the case of the Drosophila transposase, the key splicing event is blocked in somatic cells by negative regulation.
Sex determination through alternative splicing
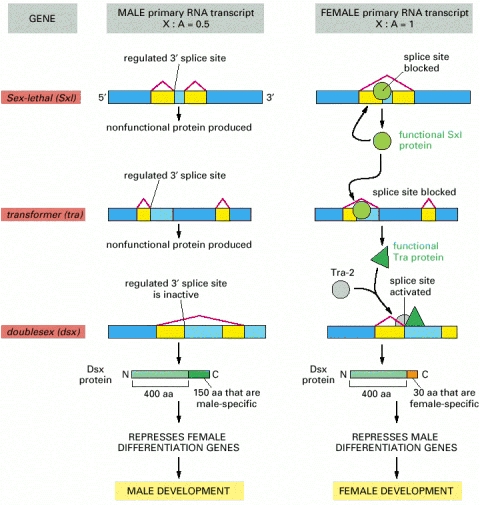
An X chromosome/ autosome ratio of 0.5 results in male development. Male is the default pathway in which the Sxl and tra genes are both transcribed, but the RNAs are spliced constitutively to produce only nonfunctional RNA molecules, and the dsx transcript is spliced to produce a protein that turns off the genes that specify female characteristics. An X chromosome/ autosome ratio of 1 triggers the female differentiation pathway in the embryo by transiently activating a promoter within the Sxl gene that causes synthesis of a special class of Sxl transcripts that are constitutively spliced to give functional Sx1 protein. Sxl is a splicing regulatory protein with two sites of action: (1) it binds to the constitutively produced Sxl RNA transcript, causing a female-specific splice that continues the production of a functional Sxl protein, and (2) it binds to the constitutively produced tra RNA and causes an alternative splice of this transcript, which now produces an active Tra regulatory protein. The Tra protein acts with the constitutively produced Tra-2 protein to produce the female-specific spliced form of the dsx transcript; this encodes the female form of the Dsx protein, which turns off the genes that specify male features. The components in this pathway were all initially identified through the study of Drosophila mutants that are altered in their sexual development. The dsx gene, for example, derives its name (doublesex) from the observation that a fly lacking this gene product expresses both male- and female-specific features. Note that, although both the Sxl and the Tra proteins bind to specific RNA sites, Sxl is a repressor that acts negatively to block a splice site, whereas the Tra proteins are activators that act positively to induce a splice. Sx1 binds to the pyrimidine-rich stretch of nucleotides that is part of the standard splicing consensus sequence and blocks access by the normal splicing factor, U2AF. Tra binds to specific RNA sequences in an exon and activates a normally suboptimal splicing signal.
Alternative splicing in B-Cells
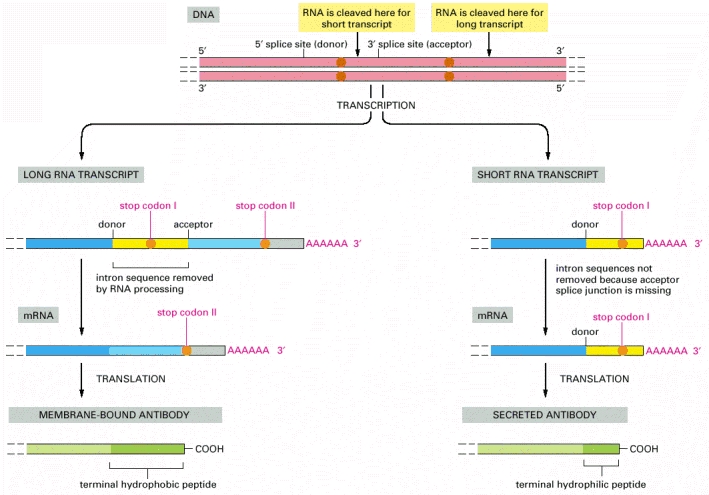
In unstimulated
B lymphocytes (left), a long RNA transcript is produced, and the intron
sequence near its 3′ end is removed by RNA splicing to give rise to an mRNA
molecule that codes for a membrane-bound antibody molecule. In contrast, after antigen
stimulation (right) the primary RNA transcript is cleaved upstream from
the splice site in front of the last exon sequence. As a result, some of the
intron sequence that is removed from the long transcript remains as coding
sequence in the short transcript. These are the nucleotide sequences that
encode the hydrophilic C-terminal portion of the secreted antibody molecule.
SPLICEOSOME ASSEMBLY
The transesterification reactions just described are mediated by a huge molecular "machine" called the spliceosome. This complex comprises about 150 proteins and 5 RNAs and is similar in size to a ribosome. In carrying out even a single splicing reaction, the spliceosome hydrolyzes several molecules of ATP. Strikingly, it is believed that many of the functions of the spliceosome are carried out by its RNA components rather than the proteins, again reminiscent of the ribosome. Thus, RNAs locate the sequence elements at the intron-exon borders and likely participate in catalysis of the splicing reaction itself.
The five RNAs (Ul, U2, U4, U5, and U6) are collectively called small nuclear RNAs (snRNAs). Each of these RNAs is between 100 and 300 nucleotides long and is complexed with several proteins. These RNA-protein complexes are called small nuclear ribonuclear proteins (snRNPspronounced "snurps"). The spliceosome is the large complex made up of these snRNPs, but the exact makeup differs at different stages of the splicing reaction: different snRNPs come and go at different times, each carrying out particular functions in the reaction. There are also many proteins within the spliceosome that are not part of the snRNPs, and others besides that are only loosely bound to the spliceosome.
The snRNPs have three roles in splicing. They recognize the 5' splice site and the branch site; they bring those sites together as required; and they catalyze (or help to catalyze) the RNA cleavage and joining reactions. To perform these functions, RNA-RNA, RNA-protein, and protein-protein interactions are all important. We start by considering some of the RNA-RNA interactions. These operate within individual snRNPs, between different snRNPs, and between snRNPs and the pre-mRNA.
Thus, for example, the interaction, through complementary base-pairing, of the Ul snRNA and the 5' splice site in the pre-mRNA. Later in the reaction, that splice site is recognized by the U6 snRNA. In another example, the branch site is recognized by the U2 snRNA. A third example, shows an interaction between U2 and U6 snRNAs. This brings the 5' splice site and the branch site together. It is these and other similar interactions, and the rearrangements they lead to, that drive the splicing reaction and contribute to its precision, as we will see a little later. Some RNA-free proteins are involved in splicing as mentioned above. One example, U2AF (U2 auxiliary factor), recognizes the polypyrimidine (Py) tract/3' splice site, and, in the initial step of the splicing reaction, helps another protein, branch-point binding protein (BBP), bind to the branch site. BBP is then displaced by the U2 snRNP, as shown in Figure 13-6d.
Other proteins involved in the splicing reaction include RNA-annealing factors, which help load snRNPs onto the mRNA, and DEAD-box helicase proteins. The latter use their ATPase activity to dissociate given RNA-RNA interactions, allowing alternative pairs to form and thereby driving the rearrangements that occur through the splicing reaction. Finally, before turning to the spliceosome mediated splicing pathway itself, we look at one further interaction.
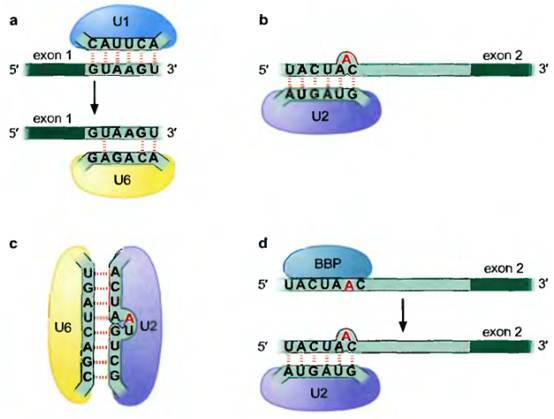
Some RNA-RNA hybrids formed during the splicing reaction. In some cases, (a) different snRNPs recognize the same (or overlapping) sequences in the pre-mRNA at different stages of the splicing reaction, as shown here for U1 and U6 recognizing the 5' splice site. In (b) snRNP U2 is shown recognizing the branch site. In (c) the RNAiRNA pairing between the snRNPs U2 and U6 is shown. Finally, in (d), the same sequence within the pre-mRNA is recognized by a protein (not part of an snRNP) at one stage and displaced by an snRNP at another. Each of these changes accompanies the arrival or departure of components of the spliceosome and a structural rearrangement that is required for the splicing reaction to proceed.
RIBOZYMES
It was widely believed for many years that only proteins could be enzymes. An enzyme must be able to bind a substrate, carry out a chemical reaction, release the product and repeat this sequence of events many times. Proteins are well-suited to this task because they are composed of many different kinds of amino acids B0) and they can fold into complex tertiary structures with binding pockets for the substrate and small molecule co-factors and an active site for catalysis.
Now we know that RNAs, which as we have seen can similarly adopt complex tertiary structures, can also be biological catalysts. Such RNA enzymes are known as ribozymes, and they exhibit many of the features of a classical enzyme, such as an active site, a binding site for a substrate, and a binding site for a co-factor, such as a metal ion. One of the first ribozymes to be discovered was RNAse P, a ribonuclease that is involved in generating tRNA molecules from larger, precursor RNAs. RNAse P is composed of both RNA and protein; however, the RNA moiety alone is the catalyst. The protein moiety of RNAse P facilitates the reaction by shielding the negative charges on the RNA so that it can bind effectively to its negatively-charged substrate. The RNA moiety is able to catalyze cleavage of the tRNA precursor in the absence of the protein if a small, positively-charged counter ion, such as the peptide spermidine, is used to shield the repulsive, negative charges. Other ribozymes carry out trans-esterification reactions involved in the removal of intervening sequences known as introns from precursors to certain mRNAs, tRNAs, and ribosomal RNAs in a process known as RNA splicing.
RNA EDITING
RNA editing, like RNA splicing, can change the sequence of an RNA after it has been transcribed. Thus the protein produced upon translation is different from that predicted from the gene sequence. There are two mechanisms that mediate editing: site-specific deamination and guide RNA-directed uridine insertion or deletion.
In one form of site-specific deamination, a specifically targeted cytosine residue within mRNA is converted into uridine by deamination. Typically, for a given mRNA species, the process occurs only in certain tissues or cell types and in a regulated manner. Figure 13-24 shows the mammalian apolipoprotein-B gene. This gene has several exons, within one of which is a particular CAA codon that is targeted for editing; it is the C within this codon that gets deaminated. That deamination, carried out by the enzyme cytidine deaminase, converts the C to a U. In this example, the deamination occurs in a tissue-specific manner: messages are edited in intestinal cells but not in liver cells. These two forms of apolipoprotein B are both involved in lipid metabolism.
The longer form, found in the liver, is involved in the transport of endogenously synthesized cholesterol and triglycerides. The smaller version, found in the intestines, is involved in the transport of dietary lipids to various tissues. Thus the CAA codon, which is translated as glutamine in the unedited message in the liver, is converted in the intestine, to UAAa stop codon. The result is that the full-length protein (of some 4,500 amino) acids is produced in the liver, but a truncated polypeptide of only about 2,100 amino acids is made in the intestine.
Other examples of mRNA editing by enzymatic deamination include adenosine deamination. This reaction carried out by the enzyme ADAR (adenosine deaminase acting on RNA)of which there are three in humansproduces Inosine. Inosine can base-pair with cytosine, and so this change can readily alter the sequence of the protein encoded by the mRNA. An ion channel expressed in mammalian brains is the target of this type of editing. A single edit in its mRNA elicits a single amino acid change in the protein, which in turn alters the Caz" permeability of the channel. In the absence of this editing, brain development is seriously impaired. A very different form of RNA editing is found in the RNA transcripts that encode proteins in the mitochondria of trypanosomes. In this case, multiple Us are inserted into specific regions of mRNAs after transcription (or, in other cases, Us may be deleted). These insertions can be so extensive that in an extreme case they amount to as many as half the nucleotides of the mature mRNA. The addition of Us to the message changes codons and reading frames, completely altering the "meaning" of the message. As an example, consider the trypanosome coxll gene. In a specific region of the mRNA of this gene, four Us are inserted between adjacent bases at three sites (two Us at one site and one U at each of two additional sites). These additions alter some codons and cause a " 1" change in the reading frame, a shift that is required to generate the correct open-reading frame.
Us are inserted into the message by so-called guide RNAs (gRNAs). These gRNAs range from 40 to 80 nucleotides in length and are encoded by genes distinct from those that encode the mRNAs they act on. Each gRNA is divided into three regions. The first, at the 5' end, is called the "anchor" and directs the gRNA to the region of the mRNA it will edit; the second determines exactly where the Us will be inserted within the edited sequence; and the third, at the 3' end, is a poly-U stretch. We now look more closely at how the gRNAs direct editing.
The anchor region of the gRNA contains a sequence that can base-pair with a region of the message immediately beside C' to) the region that will be edited. This is followed by the editing "instructions:" a stretch of gRNA complementary to the region in the message to be edited, but containing additional As. The As are at positions in the gRNA opposite where Us will be inserted into the mRNA. At the 3' end of the gRNA is the poly-U region. The role of the nucleotides in this region is unclear, though it is proposed that they tether the gRNA to purine rich sequences in the mRNA upstream E' to) the edited region.
As shown in Figure, the gRNA and mRNA form an RNA-RNA duplex with looped out single-stranded regions opposite where Us will be inserted. An endonuclease recognizes and cuts the mRNA opposite these loops. Editing involves the transfer of Us into the gap in the message. This process is catalyzed by the enzyme 3' terminal uridylyl ferase (TUTase). After the addition of Us, the two halves of the mRNA are joined by an RNA ligase, and the "editing" region of the gRNA continues its action along the mRNA in a 3' to 5' direction. A single gRNA can be responsible for inserting several Us at different sites. Furthermore, in some cases, several different gRNAs work on different regions of the same message.
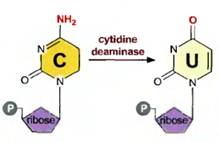
The deamination of the base cytosine to produce uracil.
RNA editing by deamination. The RNA made from the human apolipoprotein gene is edited in a tissue-specific manner by deamination of a specific cytodine to generate a uridine. This event occurs in RNAs destined for the intestine, but not those for the liver. The result, as described in the text, is that a stop codon introduced into the intestinal mRNA generates a shorter protein than that produced in the liver. The figure is not drawn to scale: thus the edited exon is exon 26; and the codon marked as filling it is in reality only a very short part of that exon.
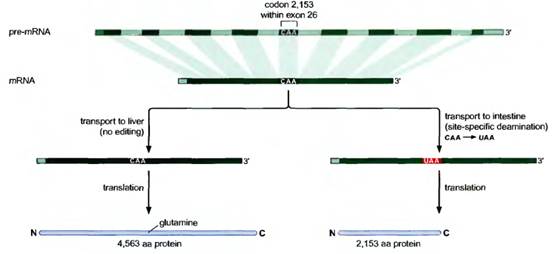
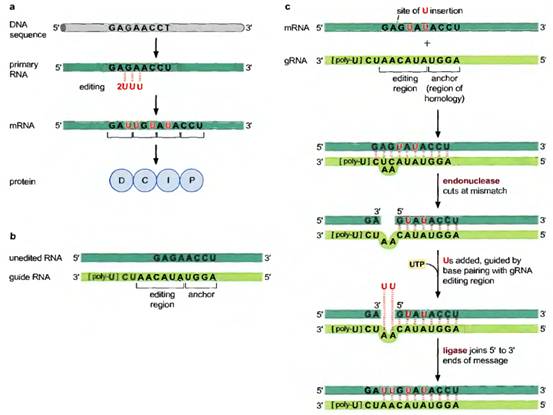
RNA editing by guide RNA mediated U insertion. Editing of the trypanosome coxll gene RNA. (a) Shows the positions of the four U nudeotides inserted into the pre-mRNA of the coxII gene These generate the correct reading frame and coding information in the mRNA. (b) Shows the sequence of the guide RNA that determines the U insertion pattern, and the sequence of the unedited stretch of mRNA. (c) Shows the editing reaction itself.
********