TRANSLATION
Translation or protein synthesis is a process in which the genetic information present in m-RNA in the form of triplet codes (codons) directs the sequence of aminoacids in protein. In otherwords, Translation is a series of events that converts the language of the genes, sequences of nucleotides into the language of polypeptides, sequence of aminoacids.
GENETIC CODE:
The set of triplet code words in m-RNA, coding for the aminoacid of protein is called genetic code. The triplet code is a codon which is the sequence of three adjacent nucleotide residues in nucleic acid that codes for a specific aminoacid. The code in form of codon is contained in m-RNA which is read by adaptor molecule (t-RNA).
Features of Genetic Code:
Genetic Code found to have the following features:
i) Genetic code is Triplet
ii) It is Non-overlapping
iii) It is Continuous
iv) It is highly Degenerate
v) It has Sense and Nonsense codons
vi) It is Almost Universal
vii) Direction of reading
i) Genetic Code is Triplet:
It is a trinucleotide sequence i.e. it is the sequence of three bases. A single base can specify only four kinds of Aminoacids because only four different types of bases available. If two bases are presumed to be the genetic cods, it can specify only 16 aminoacids [42]. If three, 64 aminoacids can be coded [43]. Since proteins are built from 20 basic aminoacids, it is evident from this simple calculation that three or more bases probably need to specify aminoacids in m-RNA.
ii) Genetic Code is Non-overlapping:
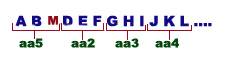
Suppose ABC, DEF, GHI and JKL are set of triplet codes specifying different aminoacids. If the genetic code is Non-overlapping, each triplet should read for only one aminoacid as shown below:

If the triplet is overlapping than ABC will code for one aminoacid, BCD will code for second aminoacid and CDE for third one and so on.

Non overlapping nature of genetic code has been proved by point mutation experiment. Had the codon being overlapping and if C is mutated then first three aminoacids will be changed because C is expressed in three codons.

But experiments with Tobacco Moasic Virus (TMV) has shown that when one base is mutated then only single aminoacid is get altered. This makes it abundantly clear that codons are Non-overlapping.
iii) Genetic Code is Continuous:
Codons are non-punctuated but are read subsequently from the fixed starting point. This means there are no commas after the reading of each triplet. Had one of the four bases say Q punctuates each triplets as shown below.
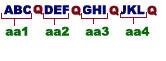
Mutation by point deletion or insertion in one of the triplet should not alter the subsequent aminoacid because each triplet is punctuated by Q base. Suppose G base is deleted it has seen that aminoacids aa1 and aa2 remains unaltered but aminoacids aa3 and aa4 get altered because the reading frame shifted leading to deletion.

In the same way, mutation by point addition also leads to altered aminoacid sequence after the point mutation site.

Thus, genetic code studies by point mutation [addition or deletion] have revealed that the codon are continuous without any punctuation and is read in a continuous fashion.
iv) Genetic Code is highly degenerate:
There are 64 possible base
triplets and 20S aminoacids. If one triplet code for one aminoacid then 20
triplet codes would have been sufficient.
Genetic studies have shown that most of the 64 triplets do code for
various aminoacids. This means that some
aminoacids are coded by more than one triplet code. The phenomena of having more than one triplet
code for the same aminoacid are called degeneracy of genetic code. For example,
v) Functional Nature of Codons:
Out of 64 codons, there are 61 functional or sense codons which specify a particular aminoacids. The remaining three, are terminating or Non-sense codons which are UAG (amber), UAA (ochre) and UGA (opal). The initiation codon is AUG and very rarely GUG.
vi) Genetic code is Almost Universal:
Analysis of mutated genes from plants, mammals, viruses and bacteria has shown that genetic code is almost universal in nature. This means that the aminoacids have mostly the same codons in all living organisms.
vii) Direction of
The genetic code triplets read in 5ΰ3 direction. Codons starting from initiation codon to until stop codons referred as open reading frame (ORF). It can be considered as single expressable gene.
DECIPHERING THE GENETIC CODE:
It is otherwise called as decoding or breaking the genetic code. Genetic code breaking made possible by M.Grunberg Manago and Ochoa. Because they provided an enzyme Polynucleotide phosphorylase which can synthesize single strand RNA by utilizing nucleoside diphoshpates. This enzyme does not require template and primer.
Genetic code deciphering achieved by the following four experiments namely:
i) Marshell Nirenberg and J.H.Mathei Experiment
ii) H.G. Korana Experiment
iii) M.Nirenberg and Philip Leader Experiment
iv) Invitro Experiment
i) M.W. Nirenberg and J.H.Mathei Experiment (1961):
This experiment was carried out with cell free extracts from bacteria. All the necessary components for protein synthesis except mRNA were present in these extracts. On the addition of chemically defined synthetic mRNAs, the extracts formed specific polypeptides.
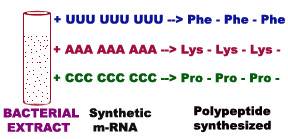
For example, synthetic m-RNA composed only of U residues yielded polypeptides made up only of phenylalanine. Likewise poly C and poly A coded for single aminoacids Proline and Lysine respectively. Poly G did not work because it assumes unstable stacked structures. Thus three codons are decoded by this experiment.
ii) H.G. Korana Experiment (1968):
When a synthetic m-RNA with alternating A and C residues was added to a protein synthesizing bacterial extract, the resulting polypeptide contained alternating threonine and histidine residues.
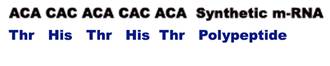
A further experiment was needed to determine whether threonine was encoded by ACA and histidine by CAC or vice versa. For this, an m-RNA consisting of repeats of AAC was tested. This m-RNA was found to stimulate the synthesis of three kinds of polypeptide chains : all aspargine, all threonine and all glutamine. This m-RNA can be read in three frames all AAC, all ACA and all CAA. Since only the ACA codon was common to both experiments, it must encode threonine. Thus CAC must encode histidine in the first experiment concluded. Comparisons of the coding capacity of many such mixed polypeptides revealed a substantial part of the genetic code.

iii) M. Nirenberg and Philip Leader Experiment:
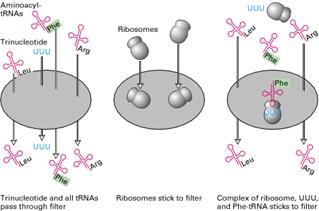
Marshall Nirenberg and his collaborators used extracts of E.Coli to which they added chemically synthesized trinucleotide to decipher the entire genetic code. They prepared 20 bacterial extracts containing all 20 aminoacyl tRNAs. In each extract sample, a different aminoacid was radioactively labeled but the other 19 aminoacids were present on tRNAs remained unlabeled. Aminoacyl tRNAs and trinucleotide passed through a nitrocellulose filter without binding whereas ribosomes did bind to the filter. Each possible trinucleotide was tested separately for its ability to attract specific tRNA by adding it with ribosome to samples from each of the 20 aminoacyl tRNA mixtures. The sample was then filtered.
If the added trinucleotide caused the radiolabeled aminoacyl tRNA to bind to the ribosome, then radioactivity would be detected on the filter. It indicated that the codon codes the corresponding aminoacid. Otherwise, the label would pass through the filter. By synthesizing and testing all possible trinucleotides, the researchers were able to match all 20 aminoacids with one or more codons.
iv) Invitro Experiment:
The first successful synthesis of a specific protein occurred when the m-RNA of bacteriophage F2 was added to bacterial extracts and the coat or capsid protein was formed. With the use of real m-RNAs, it was discovered that AUG encoded Met at the start of almost all proteins and three codons UAA, UAG and UGA did not code any aminoacid but act as terminator or stop codons.
WOBBLE HYPOTHESIS:
It is a hypothesis given by Crick to explain how one t-RNA molecule can accommodate more than one codon of aminoacids. In order to explain the above anomaly Crick proposed a word wobble which according to him is the relative loose base pairing between base at the 3-end of the codon and the complementary base at the 5-end of the anticodon in the t-RNA. The hypothesis proposes four relationships:
1. The first two bases of a codon always form strong Watson crick basepair with the corresponding bases of anticodon and confer most of the coding specificity.
2. The first base of anticodon [5ΰ3 direction] or the 3rd base [3ΰ5 direction] called the wobble base allows the single t-RNA to adopt more than one codon. The 3rd base [3ΰ5 direction] of the codon leading to loose base pairing which is termed as wobble. The wobble permits t-RNA to read more than one codon with the maximum limits of three codons.
3. If the wobble base is C or A in the anticodon, then it recognize only one codon which most contain G or U respectively in its 3 position. Here C and A form Watson-Crick base pair with G and U respectively.
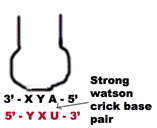
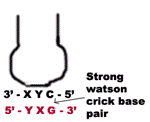
a) When the first base in the 5-end in the anticodon is U then third base in codon can be either A or G. The U forms strong Watson-Crick basepair with A and wobble pairing with G. Similarly if G present at 5 ends of anticodon, then G forms Watson-Crick basepair with C and wobble base pair with U.
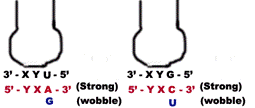
b) When I or some other modified base present at 5-end of anticodon then t-RNA can recognize three different codons, all of which form a wobble base pairing at 3 position of codon. The bases that can pair in this case are A, U and C.
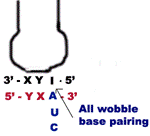
Thus part of degeneracy of the genetic code arises from wobble in the pairing of the third base of the codon.
3. For a given aminoacid and codon that differ in either of first two bases [5ΰ3 direction] requires different t-RNAs.
4. A minimum of 32 t-RNAs are required to translate all 61 different codons for the aminoacids.

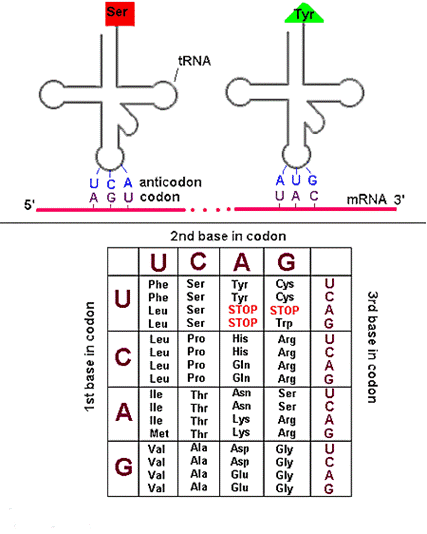
GENETIC CODE DICTIONARY:
After deciphering all codons, genetic code dictionary is formed. All 64 possible codons are listed with the amino acids they specify. The codons are read in the 5ΰ3 direction. The m-RNA Genetic code is as follows:
Prokaryotic m-RNA:
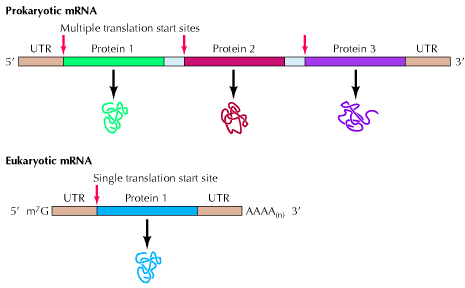
Size of m-RNA molecule is variable depending on the polypeptide chain products whose message it carries. Prokaryotic m-RNAs are polycistronic in nature i.e. they are polygenic messengers. It is a template for several polypeptide chains. For example a single m-RNA molecule codes for three specific enzymes to synthesis aminoacid tryptophan. In polycistronic m-RNA, the individual cistrons may be separated by intercistroonic sequences called spacers. The 5-end of the m-RNA may contain a sequence that is never translated into protein. These are called leader sequences or 5-untranslated region. A similar sequence is called 3-untranslated sequence may occur at the 3-end of the m-RNA. The life time of prokaryotic m-RNA is short compared to that of eukaryotic m-RNA.
In prokaryotes, the primary m-RNA transcript provides the functional m-RNA ready for translation. It has recently being found that towards the 5-end of m-RNA, there is a region of 20 or 30 nucleotides before initiation codon AUG is reached. This is nothing but the linear sequence contains the nucleotide sequence which is responsible for the interaction of the m-RNA with 30S subunit. It is known as Shine-Dalgarno (SD) sequence.

It has been showed that SD sequence binds to the complementary sequence at the 3-end of the 16S-rRNA of 30S subunit of ribosome to position the m-RNA correctly to start initiation.
Eukaryotic m-RNA:
Eukaryotic m-RNA is synthesized by RNA polymerase II. Eukaryotic m-RNAs are monocistronic in nature i.e. they are the template for the synthesis of a single polypeptide chain or it can be said that they carry codons only to code for single polypeptide chain. It has 7-methyl guanosine as a cap at its 3-end. It has poly A tail as a tail at 3-end. Exons and introns are present in it. In eukaryotes, the primary m-RNA transcript is not a functional component which is referred as heterogeneous nuclear RNA. The lifetime of eukaryotic m-RNA is greater than prokaryotic m-RNA. Kozak sequence at 5-end region of m-RNA increases the effectiveness of initiation of translation.

Ribosomes
a) Prokaryotic Ribosome 70S ribosome:
The prokaryotic ribosome is made up of two subunits 30S and 50S subunit. Ribosome contains about 65% of RNA and 35% of protein.
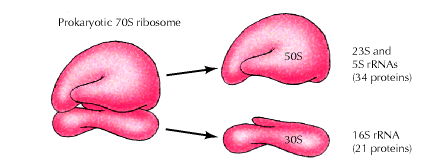
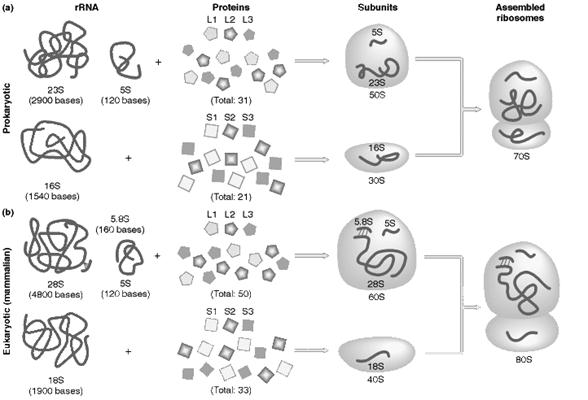
The 50S subunit comprises of 34 proteins (L-Proteins) and 23S and 5S r-RNAs. The 23S r-RNA made up of 2904 nucleotide residues and 5S r-RNA of 120 nucleotide residues. The 30S subunit consists of 21 ribosomal proteins (S-Proteins) and 16S r-RNA molecule which contain 1532 nucleotide residues. Most ribosomal proteins are low molecular weight basic protein. The basic charge reflects their ability to interact with negatively charged RNA. The RNA molecules within the ribosome have defined secondary structure and interact with the ribosomal protein in a defined manner. Prokaryotic ribosome can be dissembled into RNA and protein components and then reassembled into active functional ribosome. Ribosomal proteins are present as single copy except L-7 and L-12 proteins.
b) Eukaryotic Ribosome 80S ribosome:
It has the following structure
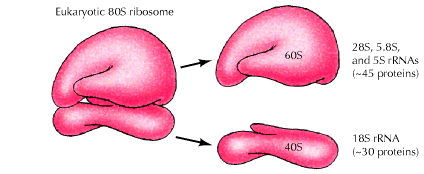
It is made up of two subunits namely large 60S subunit and smaller 40S subunit. 60S subunit contains about 40 to 45 polypeptides and 3r-RNAs components [28S r-RNA, 5.8S r-RNA and 5S r-RNA] and 40S subunit contains about 30 polypeptides and 18S r-RNA components. X-ray diffraction studies have revealed that ribosomes have the following sites
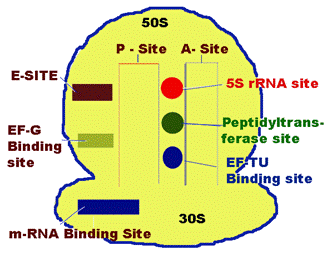
P-Site [Peptide Site]:
It is located on 30S subunit but also extent to 50S subunit. It is the site which binds to initiating t-RNA i.e. N-formyl methionine-t-RNA fmet . During translation peptide containing tRNA present in this site. So, the name Peptide site.
A-Site [Aminoacid Site]:
It lies closely to P-site. It is the site in the ribosome for the binding of incoming aminoacyl tRNA.
m-RNA binding Site:
It is located on 30S subunit. It is associated with 16S r-RNA which carries SD sequence which plays a key role in the m-RNA binding.
Peptidyl transferase site:
It lies somewhere between A and P sites. 23S r-RNA and some of the L-protens are needed for their activity.
5S r-RNA site:
It is located near peptidyl transferase site.
EF-TU binding Site:
It is also located near peptidyl transferase Site. To this site EF-TU binds during elongation of translation. It is present in 50S subunit.
EF-G binding Site:
It is located on the larger subunit of 70S ribosome close to the interface of smaller and larger subunit. To this EF-G binds during elongation of translation.
E-Site:
It is the excision site which is located on 50S subunit. Empty tRNA present at this site during translation before it is freed from ribosome.
Polysomes (Polyribosomes):
It is a complex of a messenger RNA molecule and two or more ribosomes. When ribosomes are isolated from tissues that very active in protein biosynthesis such as pancreas, they are often assayed in clusters containing several ribosomes. Such clusters of m-RNA and number of ribosomes are called polyribosomes or Polysomes. The formation of polysomes increases the efficiency of translation because the m-RNA is more efficiently used.
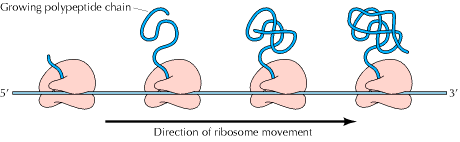
TRANSLATION IN PROKARYOTES [PROTEIN
SYNTHESIS]:
It occurs in four stages, they are
i) Activation of aminoacids
ii) Initiation of polypeptide synthesis
iii) Elongation
iv) Termination
i) Activation of aminoacids:
This involves an enzymatically catalyzed covalent binding of an aminoacid to a specific t-RNA at the 3-end at the expense of ATP. The reaction takes place in cytosol in which 20 different aminoacids are esterified to their corresponding t-RNA at their Adenosine residue at 3-end by 20 different activating enzyme called aminoacyl t-RNA synthetases, each of which is specific for one aminoacid and corresponding t-RNA. The overall reaction catalyzed by aminoacyl t-RNA synthetase is given below
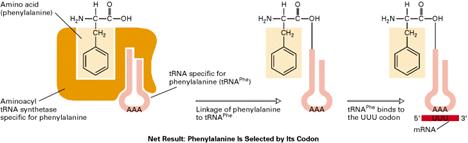
The activation occurs in two steps on enzyme catalytic site. In the first step, an enzyme bound intermediate aminoacyl adenylate is formed. In the second step, aminoacyl group is transferred from enzyme bound aminoacyl adenylate to its corresponding specific t-RNA. This reaction is irreversible.
Formylation of Methionine:
In E.coli and other prokaryotes, the starting aminoacid at the amino terminal end is always N-formylmethionine.
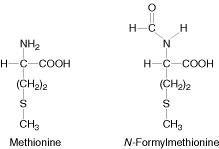
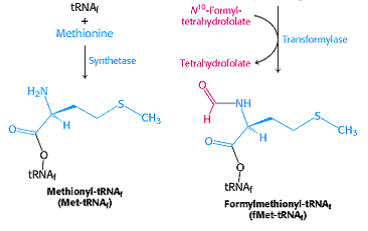
It enters as N-formyl methionyl t-RNAfmet symbolized as metf- t-RNAfmet which is formed in two successive reactions.
a) Methionine is attached to special initiating metf- t-RNAfmet by Methionyl t-RNA synthetase.
b) The formyl group is transferred to amino group of methionine residue from its donor N10-formyl tetrahydrofolate by a specific transformylase enzyme.
This enzyme cant formylate free methionine. There are two species of t-RNA specific for methionine. One is designated as t-RNAfmet and other t-RNAmet. Both can accept methionine in activation reaction but only t-RNAfmet always can accept formyl group to become the initiating aminoacid. The other species of methionine t-RNAmet is used to insert methionine in interior position in polypeptide chain.
Blocking of the amino group of methionine by N-formyl group not only prevent it from entering into interior position but also allows fmet- t-RNAfmet to be bound at the specific initiation site which does not accept methionine from other aminoacyl t-RNA.
In Eukaryotic cells, on the other hand, all polypeptides are synthesized which begins with methionine residue donated by special initiating methionyl t-RNA (t-RNAimet). Polypeptides synthesized by mitochondria ad chloroplast of eukaryotic cells begins with N-formyl Methionine.
ii) Initiation of Polypeptide Synthesis:
The formation of 70S initiation complex constitutes initiation. The conditions required for initiation are
a) m-RNA with initiation codon AUG
b) N-formyl methionyl- t-RNAfmet
c) GTP
d) 30S and 50S subunits of ribosome
e) Mg2+
f) Initiation Factors IF1, IF2, and IF3
a) Formation of IF3-IF1-30S complex:
Ribosome exists as 70S subunit in the presence of Mg2+ ions. When the Mg2+ concentration decreased, 70S subunit was dissociated into 50S and 30S subunit. The initiation factor IF3 binds with the 30S subunit so as to bring about change in shape of 30S subunit which prevents its association with 50S subunit. The other factor IF-1 also binds to smaller subunit. The IF-1 assists the binding of IF-3.
b) Binding of m-RNA:
The binding of m-RNA to 30S subunit takes place such a way that the initiating codon in m-RNA binds to the m-RNA binding site located on 30S subunit. The initiation codon AUG is guided to correct position on 30S subunit by a special signal sequence called shine dalgarno sequence (AGGAGG). Since there is only one codon for methionine which codes for both initiating and interior methionine residues, the initiation signal of SD sequence identify the site where N-formyl met-t-RNAfmet is to be bound. Interior AUG codons are specific for methionyl t-RNAmet and cannot bind N-formyl methionyl t-RNAfmet. This stage results in the formation of IF3-IF1-30S-m-RNA complex.
c) Binding of N-formyl Methionyl- t-RNAfmet:
The IF-3-IF-1-30S-m-RNA complex becomes still larger by binding to IF-2 which already contains GTP and Nfmet t-RNAfmet . During the binding, t-RNA correctly placed on initiating codon.
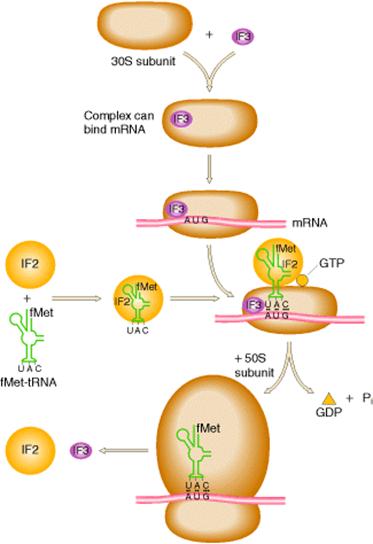
d) Formation of Initiation Complex:
In this stage, 50S ribosomal subunit combines with large complex with the simultaneous release of initiation factors IF-1, IF-2 and IF-3 which is accompanied by the hydrolysis of GTP to GDP and Pi. This gives rise to a functional initiation complex containing N-formylmethionyl t-RNAfmet m-RNA -70S ribosome.
iii) Elongation of Polypeptide Synthesis:
It occurs in three steps namely
a) Binding of aminoacyl t-RNA to A-Site
b) Peptide bond formation
c) Translocation
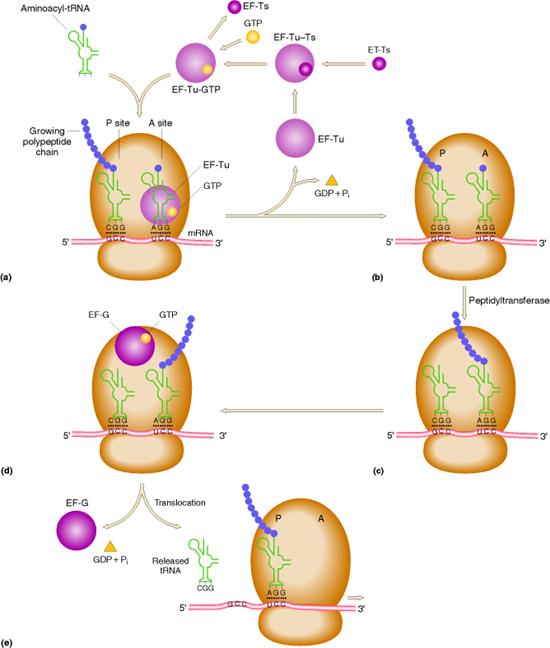
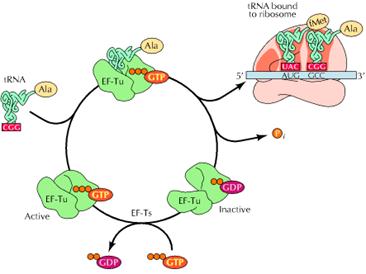
a) Binding of aminoacyl t-RNA to A Site:
In this step, the aminoacyl t-RNA is attached to the A-site with the help of elongation factors and GTP. First aminoacyl t-RNA binds with a binary complex of EF-TU-GTP to form a ternary complex. It binds to ribosome and during this reaction GTP hydrolyzed into GDP and Pi. After binding, EF-TU-GDP and Pi released. EF-TU-GDP then binds with EF-TS to form EF-TS-EF-TU-GDP complex. GTP then replaces GDP to form EF-TU-GTP along with the release of EF-TS. This EF-TU-GTP complex was now ready for the addition of another aminoacyl t-RNA to A site.
c) Peptide bond formation:
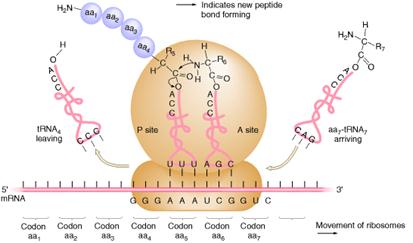
In the second step of elongation cycle a new peptide bond is formed between the aminoacids whose t-RNAs are located on A and P sites on the ribosome. This step occurs by the transfer of initiating N-formylmethionine residue from its t-RNA to amino group of the new aminoacid residue that just entered at A site. This step is catalyzed by peptidyl transferase activity provided by 23Sr-RNA and six L-proteins of 50S subunit. 5Sr-RNA and 6 other proteins also aid peptidyl transferase activity. As a result of this reaction a dipeptide if formed on t-RNA at A site and now the empty t-RNA present at P site.
d) Translocation:
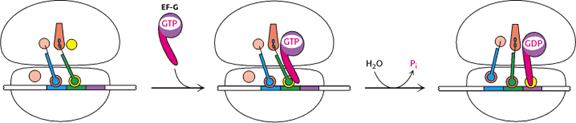
 In the third step of elongation cycle, the ribosomes move along
m-RNA towards its 3-end by the distance of one codon. Since, the dipeptidyl t-RNA is still attached
to the second codon of the m-RNA, the movements of the ribosome shifts the dipeptidyl t-RNA from the A site to P site which
causes the release of preceding t-RNAfmet which is emptied from the
P site. This shift of ribosome along
m-RNA is called translocation. This
requires elongation factor EF-G which is also called as Translocase. The hydrolysis of GTP provides energy for
Translocation.
In the third step of elongation cycle, the ribosomes move along
m-RNA towards its 3-end by the distance of one codon. Since, the dipeptidyl t-RNA is still attached
to the second codon of the m-RNA, the movements of the ribosome shifts the dipeptidyl t-RNA from the A site to P site which
causes the release of preceding t-RNAfmet which is emptied from the
P site. This shift of ribosome along
m-RNA is called translocation. This
requires elongation factor EF-G which is also called as Translocase. The hydrolysis of GTP provides energy for
Translocation.
The ribosome with its attached dipeptidyl t-RNA and m-RNA is now ready for another elongation cycle to attach the third aminoacid residue which proceeds in precisely the same way as the addition of second.
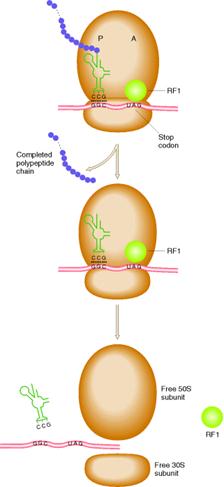
iv) Termination:
The polypeptide chain continues
to grow until one of the stop codons [UAA, UAG or UGA] is reached. These codons do not specify any aminoacid and
have no t-RNA to pair with them. When
these codons are reached, they are recognized by release factors which are one
of these types RF1, RF2 and RF3. Release
factors recognize four nucleotides in m-RNA along with the hydrolysis of
GTP. RF1 recognizes UAA and UAG. RF2 recognizes
UAA and UGA. RF3 binds with GTP, and
stimulate the binding of RF1 and RF2 to ribosomes. All these factors act at the ribosome A
site. But recently it was dentified that UAG and UGA codes for Pyrrolysine and
selenocystein respectively.
EUKARYOTIC TRANSLATION:
Eukaryotic translation occurs in four stages namely:
I) Activation of Aminoacids
II) Initiation
III) Elongation
IV) Termination
I) Activation of Aminoacids:
As in prokaryotes, aminoacids are activated by binding to t-RNA. The initiating aminoacyl t-RNA is met-t-RNAimet instead of met-t-RNAifmet.
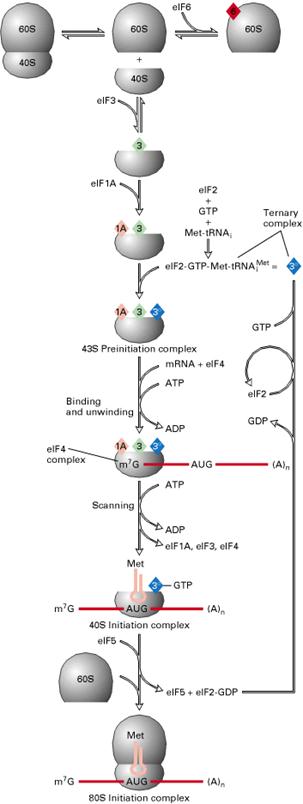
II) Initiation:
It occurs in five steps. In first step, eIF2, GTP and met-t-RNAimet bind to form ternary complex. In the second step, ternary complex bind to 40S-eIF1-eIF3-eIF4C complex to form 40S-t-RNA complex. In the third step, m-RNA-4f-4a-4b complex bound to the above complex along with the release of eIF4a, 4b and 4f. This reaction is derived by ATP hydrolysis.
In the fourth step, t-RNA recognizes initiation codon and bound to it. In the final step, 60S subunit of 80S ribosome bound along with the release of eIF1, 3, 4C and 2. eIF-5 catalyzes this step. Hydrolysis of GTP occurs in this step. Initiation complex 80S-m-RNA- met-t-RNAimet formed.
III) Elongation:
Elongation occurs in three steps as in prokaryotes namely
a) Binding of aminoacyl t-RNA to A site
b) Peptide bond formation
c) Translocation
The main difference between prokaryotes and eukaryotes is that the elongation factors. In eukaryotes the elongation factors are eEF1a, eEF1bg and eEF2.
IV) Termination:
A Single factor eRF-1 or eTF-1, was found to catalyze the release of the completed polypeptide chain from eukaryotic ribosomes. This appears to recognize all three termination codons UAA, UAG and UGA. GTP hydrolysis is required for termination.
INHIBITORS: PROKARYOTIC
INHIBITORS:
|
S.No |
Name |
Mechanism of Action |
|
1. |
Streptomycin, Neomycin and Kanamycin |
Binds to 30S subunit to cause misreading and inhibition of initiation. |
|
2. |
Paromomycin |
Inhibit initiation. Resistant mitochondria have altered small r-RNA. |
|
3. |
Tetracycline |
Inhibits elongation by blocking binding of aminoacyl t-RNA to the A site on the 30S subunit. |
|
4. |
Chloramphenicol |
Inhibits elongation at Peptidyl transferease activity. |
|
5. |
Erythromycin |
Inhibits elongation at Transpeptidation step. |
|
6. |
Spectinoycin |
Inhibits elongation at transpeptidation. Resistant ribosomes have altered potein S5. |
|
7. |
Thiostrepton |
Inhibits elongation, preventing binding of EF-G-GTP complex to ribosome. |
|
8. |
Kirromycin |
Inhibits elongation, preventing release of EF-TU-GDP complex from ribosome. |
|
9. |
Colicin E3 |
Specific nuclease for site on 16S r-RNA. |
|
10 |
Trimethoprin |
Prevents formation of fmet-t-RNA by inhibition of synthesis of N10-formyl THF. |
|
11. |
Linomycin |
Inhibit peptidyl transferase complex |
|
12. |
Kasugamycin |
Inhibit binding of aminoacylt-RNAfmet . |
EUKARYOTIC INHIBITORS:
|
S.No |
Name |
Mechanism of Action |
|
1. |
Cycloheximide (actidione) |
Inhibits elongation and initiation, freezing ribosomes on polysomes (peptidyl transferase). |
|
2. |
Emetine |
Inhibits elongation at translocation step. Resistant hamster cells have altered protein S14. |
|
3. |
Diphtheria toxin |
Inhibits elongation by inactivating EF-2 by ADP ribosylation. |
|
4. |
Ricin and abrin |
Inhibits elongation, affecting 60S subunit. |
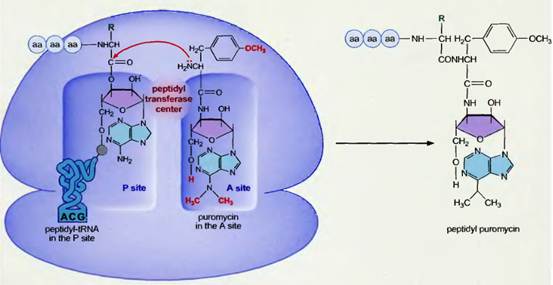
BOTH:
|
S.No |
Name |
Mechanism of Action |
|
1. |
Puromycin |
Inhibits elongation due to premature termination by acting as analogue of charged t-RNA. |
|
2. |
Pactamycin |
Inhibits initiation. It was used to determine gene order of proteins derived from picornaviral polyproteins. |
|
3. |
Aurintricarboxylic acid |
Inhibits initiation by preventing binding of m-RNA to ribosome. |
|
4. |
Showdomycin |
Inhibits initiation at the stage of ternary complex formation [IF2-t-RNA-GTP]. |
|
5. |
Sparsomycin |
Inhibits elongation at Peptidyl transferase step. |
|
6. |
Fusidic acid |
Inhibits elongation, preventing release of EF-G-GDP complex from ribosome. Resistant bacteria have altered EF-G. |
|
7. |
a-Sarcin |
Specific nuclease for site on 23S and 28S r-RNA in ribosomes although inactive against intact E.Coli. |
|
8. |
5-flurotryptophan |
Prevents activation of t-RNAtrp by competitive inhibition of synthetase. |
|
9. |
Norvaline |
Prevents activation of t-RNAval. |
|
10. |
Ethionine |
Causes synthesis of abnormal proteins with ehionine instead of methionine. |
|
11. |
O-Methylthreonine |
Causes synthesis of abnormal proteins with O-methylthreonine replacing threonine. |
REGULATION OF TRANSLATION
PROKARYOTIC TRANSLATIONAL CONTROL
In prokaryotes, translational control is of lesser importance than transcriptional control for two reasons. First, messenger RNAs are extremely unstable, so there is no need to control rate of translation because m-RNA lysed immediately. Second, although there are some indications of translational control in prokaryotes, such control is inefficient because energy is wasted in synthesizing m-RNAs that may never be used. Different ways in which prokaryotic translation controlled are as follows:
a) Gene location
b) Antisense RNA
c) Efficiency of m-RNA to bind with ribosome
d) Codon preference
e) Stringent Response
f) Attenuation
a) Gene Location:
Translation rate ratio of genes in Lac operon from 5-end, is 10:5:2. The reason for this is that in prokaryotes transcription and translation occur hand by hand so, genes at the beginning of operon [5-end] translated well before, the genes at the 3-end. In addition to this, exonuclease seems to degrade m-RNA more efficiently from the 3-end. Thus, genes located at 5-end of operon found to have higher translation rate than at 3-end. Usually, biologically active genes present at 5-end.
b) Antisense RNA:
Translation can also be regulated by RNA-RNA hybridization. RNA complementary to the 5-end of a m-RNA can prevent the translation of that messenger RNA. Several examples of this type of regulation are known. The regulating RNA is called antisense RNA. It is synthesized a direction which is opposite to the direction of m-RNA. Example: The m-RNA from the omp-F gene in E.Coli is prevented from being translated by complementary base pair binding with an antisense RNA called mic F-RNA.
c) Efficiency of m-RNA to bind with ribosome:
A third translational control mechanism consists of the efficiency with which the m-RNA is bound to the ribosome. This efficiency is related to some extent to the sequence of nucleotides at the 5-end of the messenger RNA that is complementary to the 3end of the 16S r-RNA in ribosome i.e. Shine Dalgarno sequence. Variations from this consensus sequences have different efficiencies of binding and therefore, the initiation of translation occurs at different rates.
d) Codon Preference:
Even though degeneracy present for aminoacids, in different species, specific codons are preferred to code for particular aminoacids. This is referred as codon preference. Thus, genes with preferable codons expressed at higher rate whereas ones with other codons expressed at slower rate.
e) Stringent Response [Idling reaction]:
Refer from Transcriptional Regulation
f) Attenuation:
Refer from Transcriptional Regulation
EUKARYOTIC TRANSLATION CONTROL:
Eukaryotic m-RNAs are much longer lived than prokaryotic ones, so there is more opportunity for translational control. The rate limiting step in translation is usually initiation, so mostly control exerted at this level. Regulation achieved mainly in two ways. They are:
i) phosphorylation
ii) Interaction by RNA binding Protein
i) Phosphorylation:
Phosphorylation of some of the regulatory factors inhibit translation whereas in some other cases it ca be stimulatory.
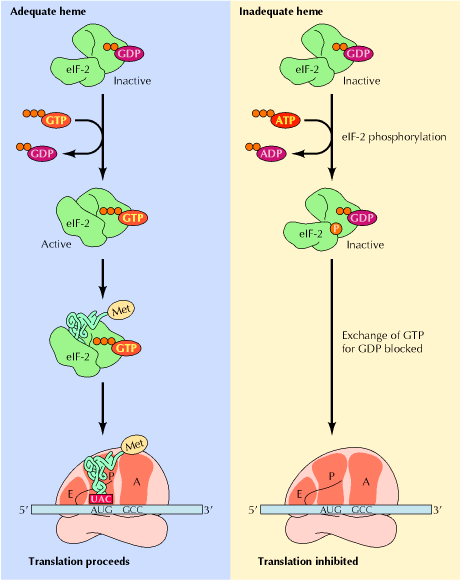
a) Inhibitory phosphorylation:
The best known example of inhibitory phosphorylation occurs in reticulocytes where in the absence of Heme, Heme controlled repressor (HCR) or Heme regulated inhibitor (HRI) phosphorylates one of the subunits of eIF2, known as eIF2a . The phosphorylated form of eIF2 binds more tightly than usual to eIF2B, which is an initiation factor whose job it is to exchange GTP for GDP on eIF2. When eIF2B is stuck fast to phosphorylated eIF2; it cannot get free to exchange GTP for GDP on other molecules of eIF2, so eIF2 remains in the inactive GDP-bound form and cannot attach met-tRNAimet to 40S ribosomes. Thus, translation initiation inhibited.
b) Stimulatory phosphorylation:
Insulin or growth factors such as EGF bind to its receptors at the cell surface. Through a series of steps these activates the signal transducer Ras, which through another series of steps, activates MAP kinase. This kinase has many targets, one of which is PHAS-1. When PHAS-1 is phosphorylated by MAP kinase, it dissociates from cap binding protein eIF4f. Now eIF4f, free to participate in translation. Thus phosphorylation stimulates translation.
ii) Interaction with RNA binding Protein:
RNA binding proteins by its interaction with RNA, it may stimulate or inhibit translation. Examples for this are as follows:
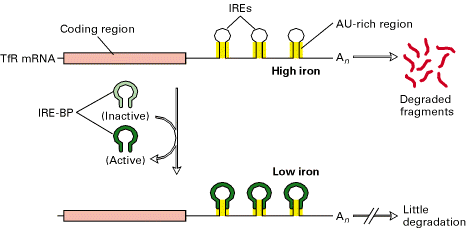
a) Stimulation of Translation:
The 3-Iron Response Element (IREs) in the transferring receptor (TfR) m-RNA have a stem loop structure containing AU rich sequence that promotes m-RNA degradation at high intracellular iron concentrations. At low intracellular iron concentrations, the concentration of IRE-BP is such that it binds to the IREs, thereby inhibiting degradation. As a result, the translation of TfR m-RNA increased because of IRE-BP.
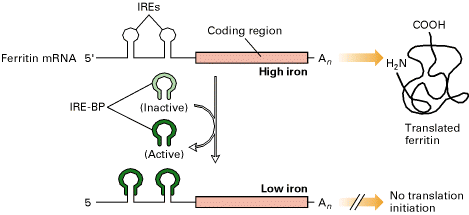
b) Inhibition of Translation:
Ferritin m-RNA contain IRE at
5-end. At low iron concentrations,
IRE-BP binds to IREs and inhibit translation of ferritin mRNA. The same mechanism controls translation of
the mRNA encoding
POST TRANSLATIONAL MODIFICATIONS:
The newly synthesized polypeptide chain by the ribosome often doesnt attain its final biologically active conformation until it has been subjected to processing of covalent modification of polypeptide chain or protein after the biosynthesis, so that they are rendered biologically active, is called post translational processing or post ribosomal modification. Several kinds of processing may takes place depending upon the nature of protein and they are as follows: I. Covalent modification and II. Non-covalent modification
I. Covalent modification:
1. Covalent modification by charge removal:
This is done by amino terminal and carboxy terminal modification in which the charge of the amino acid and carboxy terminal of the protein are removed. The amino terminal is modified by acetylation.
![]()
The carboxy terminal is modified by converting to amides.
![]()
Gastrins (hormones) are complexes of polypeptide which modified by the acetylation of their amino terminal amino group. Modification of terminal carboxy group by terminal amidation takes place in several small peptide hormones. For example: thyrotropin releasing factor in which the carboxy terminal of proline of tripeptide is amidated.
2. Covalent modification by adding negative charges:
This is done by mainly by phosphorylation which is reversible and one of the most common post translational modifications. In phosphorylation the amino acids which are most commonly phosphorylated are serine, threonine and tyrosine, lysine, arginine, and histidine group of protein are also phosphorylated, but are less common. These aminoacids are enzymatically phosphorylated by ATP.
Significance of post translational modification by phosphorylation:
There are many regulatory enzymes which plays crucial roles in the metabolism of that are covalently modified by phosphorylation. This being a reversible modification helps in the interconversion of enzyme from active to inactive form. Important example is the enzyme glycogen phosphorylase of muscle and liver which catalyzes the following reaction
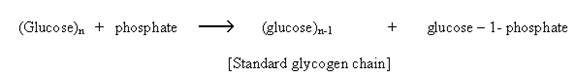
The enzyme glycogen phosphorylase exists in two forms: glycogen phosphorylase b relatively active form and glycogen phosphorylase a more active form.
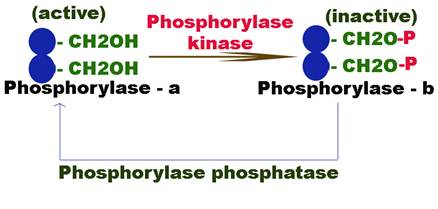
The inactive phosphorylase b is converted to active phosphorylase a by an enzyme phosphorylase kinase which phosphorylates the OH group of the serine. The phosphorylation of serine residue makes the enzyme active because they happened to be situated in their active centre i.e. catalytic site.
The active form of the enzyme is again converted back to its inactive form b by another enzyme phosphorylase phosphatase.
Phosphorylation of some protein controls the cell cycle. Phosphorylation also modulates the activity of many peptide hormones. Milk protein casein is also phosphorylated to its serine residue due to which it gets bound to calcium ions. They help in providing calcium as a nutrient phosphorylation of specific tyrosine residue has been found to be an important step in the transformation of normal cells into cancer cells.
3. Covalent modification by ADP ribosylation:
Some proteins get modified by
adding ribose to carboxylic group in proteins.
The ADP-ribose is donated by NAD+.
For example, the eEF2 gets modified by addition of ribose to the carboxy
group of histidine, when infected with Cornebacterium
diptheriae.
4. Covalent modification by methylation:
Several proteins are modified by methylation some protein lysine residues are methylated enzymematically. For example: In some muscle cell protein and cytochrome, the lysine residue and structural function. Lysine residue of some nucleoproteins may have 1-3 methyl groups.
Methylation of aspartate residue occurs in membrane protein. Methylation of either glutamate or aspartate residue destroys the negative charge. The methyl esters are readily hydrolyzed to regenerate the original carboxy groups. So, methylation of this type can be used as reversible control devices. In some of muscle protein histidine residue are methylated.
In some proteins the arginine residue are methylated especially in cell nuclei, nerve sheaths and sperm tails, sperm immobility, a common cause of human infertility is often caused by defective metabolism of sperm protein.
5. Covalent modification of hydroxylation:
In protein collagen, proline residues are hydroxylated to hydroxyproline in endoplasmic reticulum, reaction that increases the strength of collagen.
6. Covalent modification by carboxylation:
In some protein an extra carboxyl group is added to aspartic acid and glutamic acid residues. For example in blood clotting protein prothrombin a number of glutamic acid residues are carboxylated to form carboxy glutamate which increases its affinity for calcium ions. Thus it helps in initiating blood clotting.
7. Covalent Modification by glycosylation:
Many proteins that function excellulary, secretary protein ment for extracellular transport are glycosylated. For example, some of the digestive enzyme peptide hormones, membrane proteins, antibodies are glycosylated. The carbohydrate is linked mostly to aspargine and more rarely to serine or threonine residue.
Modification of glycosylation occurs both in endoplasmic reticulum and Golgi apparatus. Glycosylated involves addition of oligosaccharide consisting of N-acetyl glucosamine, mannose and galactose. Further fucose and N-acetyl neuraminic acid (NANA) are added in Golgi apparatus. Glycosylation helps in targeting and protection of proteins. For example thyroglobulin secreted by thyroid cells is glycosylated. Many proteins that lubricate mucous membrane are also glycosylated.
8. Covalent modification by addition of prosthetic group:
Many enzymes contain covalently bound prosthetic group necessity for this activity. These are attached to the polypeptide chain after it cleaves the ribosome. For example biotin molecules [prosthetic group] are covalently bound to acetyl CoA carboxylase and heam group to cytochrome C.
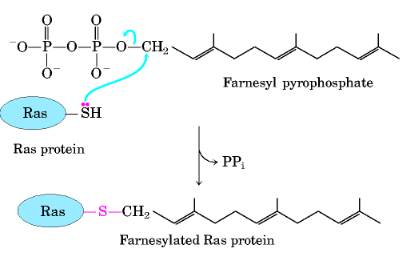
Addition of lsoprenyl Groups A number of eukaryotic proteins are modified by the addition of groups derived from isoprene (isoprenyl groups). A thioether bond is formed between the isoprenyl group and a Cys residue of the protein. The isoprenyl groups are derived from pyrophosphorylated intermediates of the cholesterol biosynthetic pathway, such as farnesyl pyrophosphate. Proteins modified in this way include the Ras proteins, products of the ras oncogenes and proto-oncogenes, and G proteins and lamins, proteins found in the nuclear matrix. The isoprenyl group helps to anchor the protein in a membrane. The transforming (carcinogenic) activity of the ras oncogene is lost when isoprenylation of the Ras protein is blocked, a finding that has stimulated interest in identifying inhibitors of this posttranslational modification pathway for use in cancer chemotherapy.
II) Non-covalent modification:
They include following:
1. Partial hydrolysis:
Enzymes involved in digestion and proteins involved clotting and some peptide hormones are modified by this method. Digestive enzymes are usually produced in their native form called zymogen because they may prove to be fatal; if they are in their active form within the cells therefore enzyme reaching the digestive track under set conditions gets converted into their active form. This modification involves partial hydrolysis of extra polypeptide chain. For example, pepsinogen, prothrombin, proinsulin get converted to their active form by partial hydrolysis.
2. Disulphide crosslink formation:
Many proteins after their biosynthesis form disulphide linkage so that they may attain a conformation to be physiologically active. For example insulin and ribonuclease enzyme require disulphide bridge formation for their catalytic activity. The disulphide bridges are formed between cysteine and methionine residue.
3. Addition of signal sequence and their removal:
Some newly made protein will simply be delivered into the cell cytosol. Some will deliver to different cell organelles. Some will be secreted into the exterior of the cell and some will be inserted into the cell membrane. It is therefore important that synthesized protein find its way to its correct site in the cell. This is done by the addition of signal sequence to the protein.
Many proteins contain specific polypeptide leaders on their amino terminal end that function as signals spot direction then to the proper destination. They are called signaling sequence, which have 15-30 aminoacid residues many of which are hydrophobic R-groups. The proteins which have to be exported out have to be targeted to Golgi bodies from where they are secreted out. Thereafter signal sequence is removed from the protein by the introns of special peptides.
The protein which are meant for organelle such mitochondria and after reaching the organelles, the signal sequence hence help in the transport of massive protein across the organelle membrane.
PROTEIN SORTING
Firefly luciferase, a peroxisomal matrix protein, is transported to peroxisomes of normal human fibroblasts, but remains cytoplasmic in cells from a Zellweger syndrome patient. The fibroblasts (on coverslips) were microinjected with mRNA encoding the luciferase. After overnight incubation in a humidified CO2 incubator, the cells were fixed, permeabilized, and labeled with appropriate primary (rabbit anti-luciferase) and secondary (FITC anti-rabbit) antibodies. The punctate immunofluorescence observed in normal human HS68 cells is indicative of peroxisomal luciferase. The fibrobrast cell line (GM6231) from the human patient does not import luciferase into peroxisomes, but shows a cytoplasmic signal instead of the punctate signal.
A typical mammalian cell contains up to 10,000 different kinds of proteins; a yeast cell, about 5000. For a cell to function properly, each of its numerous proteins must be localized to the correct cellular membrane or aqueous compartment (e.g., the mitochondrial matrix, chloroplast stroma, lysosomal lumen, or cytosol). Hormone receptor proteins, for example, must be delivered to the plasma membrane if the cell is to recognize hormones, and specific ion-channel and transporter proteins are needed if the cell is to import or export the corresponding ions and small molecules. Water-soluble enzymes such as RNA and DNA polymerases must be targeted to the nucleus; still others, such as proteolytic enzymes or catalase, must go to lysosomes or peroxisomes, respectively. Many proteins, such as hormones and components of the extracellular matrix, must be directed to the cell surface and secreted.
The process of directing each newly made polypeptide to a particular destination referred to as protein targeting, or protein sorting is critical to the organization and functioning of eukaryotic cells. This process occurs at several levels. Few proteins, encoded by the DNA present in mitochondria and chloroplasts, are synthesized on ribosomes in these organelles and are incorporated directly into compartments within these organelles. However, most mitochondrial and chloroplast proteins and all of the proteins in the other organelles, particles, and membranes of a eukaryotic cell are encoded by nuclear DNA, are synthesized on ribosomes in the cytosol, and are distributed to their correct destinations via the sequential action of several sorting signals and multiple sorting events. How nuclear-encoded organelle, membrane, and secretory proteins are sorted to their correct destinations are the major subjects of this chapter.
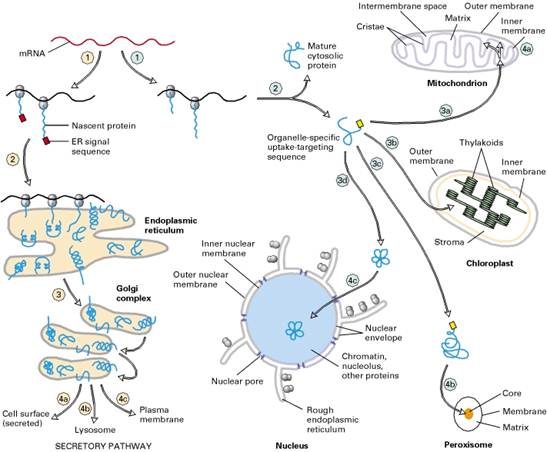
Overview of sorting of nuclear-encoded proteins in
eukaryotic cells. All nuclear-encoded mRNAs are translated on cytosolic ribosomes.
Ribosomes synthesizing nascent proteins in the secretory pathway 1 are directed to the rough
endoplasmic reticulum (ER) by an ER signal sequence 2 . After translation is completed
in the ER, these proteins move via transport vesicles to the Golgi complex dlccirc3; from whence they are further sorted to several destinations 4a, 4b, 4c . After synthesis of proteins lacking an ER signal
sequence is completed on free ribosomes 1 , the proteins are released into the cytosol 2 . Those with an organelle- specific uptake-targeting
sequence are imported into the mitochondrion 3a , chloroplast 3b , peroxisome 3c , or nucleus 3d . Mitochondrial and chloroplast proteins typically
pass through the outer and inner membranes to enter the matrix or stromal
space, respectively. Some remain there, and some 4a are sorted to other organellar
compartments. Unlike mitochondrial and chloroplast proteins, which are imported
in a partially unfolded form, most peroxisomal proteins cross the peroxisome
membrane as fully folded proteins 4b .
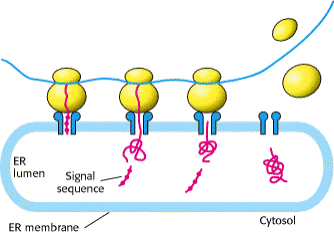
The first sorting event occurs during initial growth of nascent polypeptide chains on cytosolic ribosomes. Some nascent proteins contain, generally at the amino terminus, a specific signal, or targeting, sequence that directs the ribosomes synthesizing them to the endoplasmic reticulum (ER). Protein synthesis is completed on ribosomes attached to the rough ER membrane (the presence of these bound ribosomes distinguishes the rough ER from the smooth ER). The completed polypeptide chains then move to the Golgi complex and subsequently are sorted to various destinations. Proteins synthesized and sorted in this pathway, referred to as the secretory pathway, include not only those that are secreted from the cell but also enzymes and other resident proteins in the lumen of the ER, Golgi, and lysosomes as well as integral proteins in the membranes of these organelles and the plasma membrane.
Synthesis of all other nuclear-encoded proteins is completed on free cytosolic ribosomes, and the completed proteins are released into the cytosol. These proteins remain in the cytosol unless they contain a specific signal sequence that directs them to the mitochondrion, chloroplast, peroxisome, or nucleus. Many of these proteins are subsequently sorted further to reach their correct destinations within these organelles; such sorting events depend on yet other signal sequences within the protein. Each sorting event involves binding of a signal sequence to one or more receptor proteins on the surface or interior of the organelle.
In this chapter, we detail the mechanisms whereby proteins are sorted to the major organelles and compartments of the cell. The first two sections cover targeting of proteins to mitochondria, chloroplasts, and peroxisomes. The next several sections describe the various components and events in the secretory pathway, including the post-translational modifications that occur to proteins as they move through this pathway. We then discuss how proteins are internalized into cells following binding to specific cell-surface receptors and the fate of such internalized proteins. In the final section, we describe how the various small membrane-bounded vesicles that carry proteins within cells are formed and deliver their contents to specific destinations.
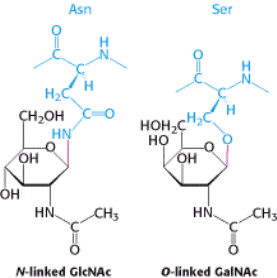
Glycosidic Bonds between Proteins and Carbohydrates. A glycosidic bond
links a carbohydrate to the side chain of asparagine (N-linked) or to
the side chain of serine or threonine (O-linked). The glycosidic bonds are shown in red.
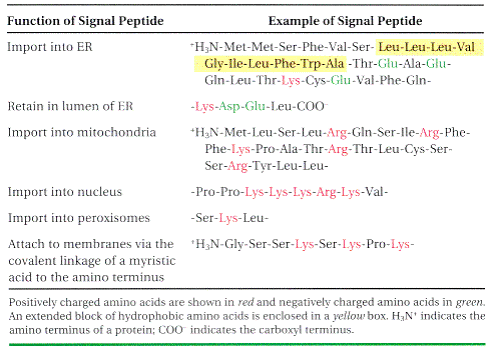
SIGNAL SEQUENCES
The most important element in many of these targeting pathways is a short sequence of amino acids called a signal sequence, whose function was first postulated by Giinter Blobel and colleagues in 1970. The signal sequence directs a protein to its appropriate location in the cell and, for many proteins, is removed during transport or after the protein has reached its final destination. In proteins slated for transport into mitochondria, chloroplasts, or the ER, the signal sequence Giinter Blobel George Ralade is at the amino terminus of a newly synthesized polypeptide. In many cases, the targeting capacity of particular signal sequences has been confirmed by fusing the signal sequence from one protein to a second protein and showing that the signal directs the second protein to the location where the first protein is normally found. The selective degradation of proteins no longer needed by the cell also relies largely on a set of molecular signals embedded in each protein's structure.
PROTEIN GLYCOSYLATION AT ER
Protein glycosylation takes place inside the lumen of the endoplasmic reticulum (ER) and the Golgi complex, organelles that play central roles in protein trafficking. One such glycoprotein is the proteolytic enzyme elastase, which is secreted by the pancreas as a zymogen. This protein is synthesized by ribosomes attached to the cytoplasmic face of the ER membrane, and the peptide chain is inserted into the lumen of the ER as it grows, guided by a signal sequence of 29 amino acids at the amino terminus. This signal sequence, which directs the protein through a channel in the ER membrane, is cleaved from the protein in the transport process into the ER. After the protein has entered the ER, the glycosylation process begins. The N-linked glycosylation begins in the ER and continues in the Golgi complex, whereas the O-linked glycosylation takes place exclusively in the Golgi complex.
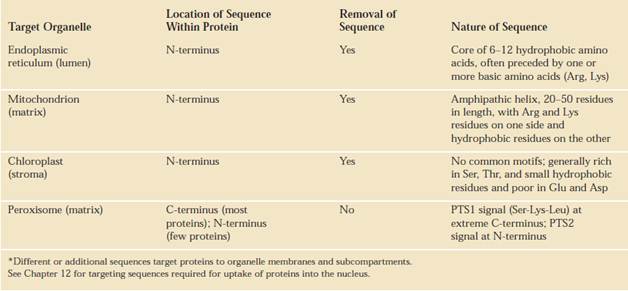
Signal sequences vary in length from 13 to 36 amino acid residues, but all have the following features: A) about 10 to 15 hydrophobic amino acid residues; B) one or more positively charged residues, usually near the amino terminus, preceding the hydrophobic sequence; and C) a short sequence at the carboxyl terminus (near the cleavage site) that is relatively polar, typically having amino acid residues with short side chains (especially Ala) at the positions closest to the cleavage site.
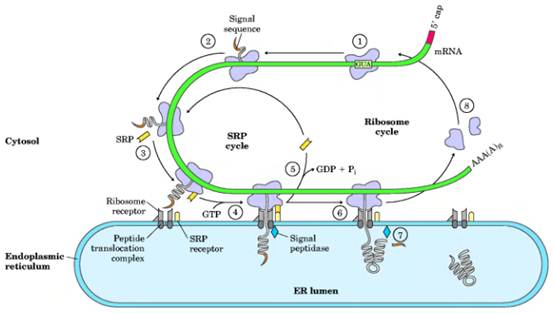
Translocation of peptide to ER explained in the following steps: 1 The targeting pathway begins with initiation of protein synthesis on free ribosomes. 2) The signal sequence appears early in the synthetic process, because it is at the amino terminus, which as we have seen is synthesized first. 3) As it emerges from the ribosome, the signal sequenceand the ribosome itselfare bound by the large signal recognition particle (SRP); SRP then binds GTP and halts elongation of the polypeptide when it is about 70 amino acids long and the signal sequence has completely emerged from the ribosome. 4) The GTP-bound SRP now directs the ribosome (still bound to the mRNA) and the incomplete polypeptide to GTP-bound SRP receptors in the cytosolic face of the ER; the nascent polypeptide is delivered to a peptide translocation complex in the ER, which may interact directly with the ribosome. 5) SRP dissociates from the ribosome, accompanied by hydrolysis of GTP in both SRP and the SRP receptor. 6) Elongation of the polypeptide now resumes, with the ATP-driven translocation complex feeding the growing polypeptide into the ER lumen until the complete protein has been synthesized. 7) The signal sequence is removed by a signal peptidase within the ER lumen; 8) the ribosome dissociates and is recycled.
IMPORTATN EVENTS AT ER
- Glycosylation occurs in ER
- Disulphide bond formation in ER
- Proper folding and assembly of multisubunit protein in ER
- Specific protein cleavage occurs in ER
GLYCOSYLATION
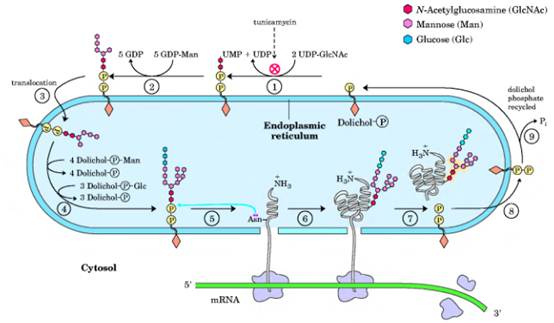
Synthesis of the core oligosaccharide of glycoproteins. The core oligosaccharide is built up by the successive addition of monosaccharide units. (1), (2) The first steps occur on the cytosolic face of the ER. 3) Translocation moves the incomplete oligosaccharide across the membrane (mechanism not shown), and 4) completion of the core oligosaccharide occurs within the lumen of the ER. The precursors that contribute additional mannose and glucose residues to the growing oligosaccharide in the lumen are dolichol phosphate derivatives. In the first step in the construction of the N-linked oligosaccharide moiety of a glycoprotein, 5, 6 the core oligosaccharide is transferred from dolichol phosphate to an Asn residue of the protein within the ER lumen. The core oligosaccharide is then further modified in the ER and the Golgi complex in pathways that differ for different proteins. The five sugar residues shown surrounded by a beige screen (after step 7)) are retained in the final structure of all /V-linked oligosaccharides. 8) The released dolichol pyrophosphate is again translocated so that the pyrophosphate is on the cytosolic face of the ER, then 9) a phosphate is hydrolytically removed to regenerate dolichol phosphate.
DISULPHIDE BOND FORMATION
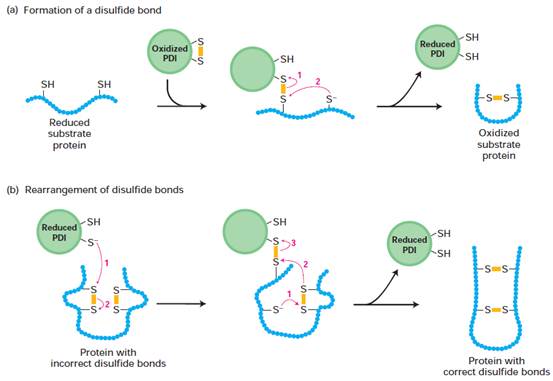
Formation and rearrangement of
disulfide bonds by protein disulfide isomerase (PDI). PDI contains an active site with two closely
spaced cysteine residues that are easily interconverted between the reduced
dithiol form and the oxidized disulfide form. Numbered red arrows indicate the
sequence of electron transfers. Yellow bars represent disulfide bonds. (a) In
the formation of disulfide bonds, the ionized (S--) form of a
cysteine thiol in the substrate protein reacts with the disulfide (S-S) bond in
oxidized PDI to form a disulfide-bonded PDIsubstrate protein intermediate. A
second ionized thiol in the substrate then reacts with this intermediate,
forming a disulfide bond within the substrate protein and releasing reduced
PDI. (b) Reduced PDI can catalyze rearrangement of improperly formed disulfide
bonds by similar thiol-disulfide transfer reactions. In this case, reduced PDI
both initiates and is regenerated in the reaction pathway. These reactions are
repeated until the most stable conformation of the protein is achieved.
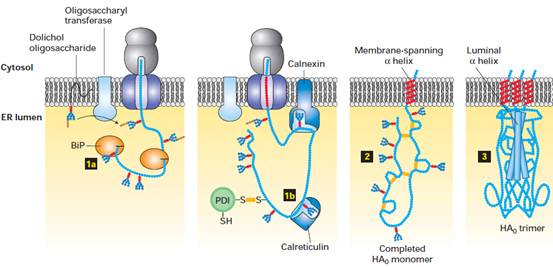
FOLDING AND ASSEMBLY AT ER
Folding and assembly of hemagglutinin
(HA0) trimer in the ER. Transient binding of the chaperone BiP (step1A ) to the nascent chain
and of two lectins, calnexin and calreticulin, to certain oligosaccharide
chains (step 1B) promotes proper folding of adjacent segments. A total of seven
N-linked oligosaccharide chains are added to the luminal portion of the
nascent chain during cotranslational translocation,
and PDI catalyzes the formation of six disulfide bonds per monomer. Completed
HA0 monomers are anchored in the membrane by a single membrane-spanning a helix with their N-terminus in the lumen (step2 ). Interaction of three HA0 chains with one another,
initially via their transmembrane a helices,
apparently triggers formation of a long stem containing one a helix from the luminal part of each HA0 polypeptide.
Finally, interactions between the three globular heads occur, generating a
stable HA0 trimer (step3).
DEGRADATION OF MISFOLDED PROTEIN
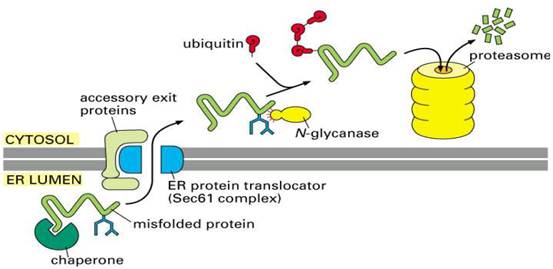
If proper folding and assembly fail to occur, misfolded
proteins may be removed by cellular quality control mechanisms. The ER-Associated
Degradation (ERAD) here involves translocation to the cytosol (retrotranslocation, through a modified translocation
complex, associated with exit proteins), followed by ubiquitination and proteasomal
degradation in the cytosol.
ROLE OF THE KDEL
RECEPTOR IN THE RETRIEVAL OF ER-RESIDENT PROTEINS
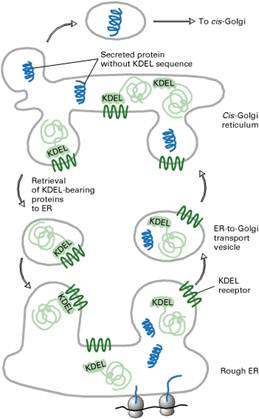
Many resident proteins in the ER lumen bear a C-terminal KDEL (Lys-Asp-Glu-Leu) sequence that localizes them to the ER. The KDEL receptor is located mainly in the cis-Golgi network and in ER-to-Golgi transport vesicles; its chief function is to bind proteins with the KDEL recognition sequence and return them to the ER.
PROTEIN SORTING AT GOLGI COMPLEX
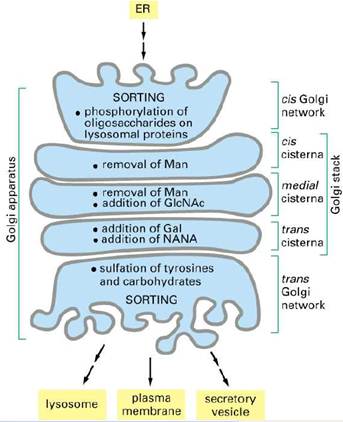
Proteins in the lumen of the ER and in
the ER membrane are transported to the Golgi complex,
which is a stack of flattened membranous sacs. The Golgi complex has two
principal roles. First, carbohydrate units of glycoproteins are altered and
elaborated in the Golgi complex. The O-linked sugar units are
fashioned there, and the N-linked sugars, arriving from the ER as a
component of a glycoprotein, are modified in many different ways. Second, the
Golgi complex is the major sorting center of the cell. Proteins proceed
from the Golgi complex to lysosomes, secretory granules (as is the case for the
elastase zymogen), or the plasma membrane, according to signals encoded within
their amino acid sequences and three-dimensional structures.
PROCESSING
OF N-LINKED OLIGOSACCHARIDES IN THE GOLGI
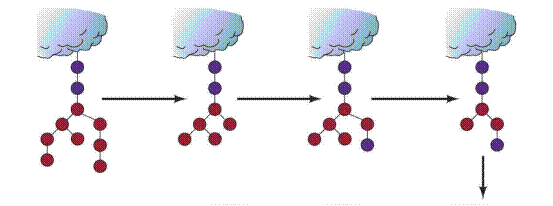
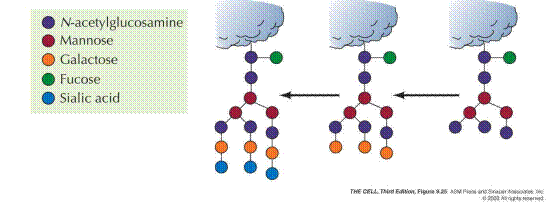
The Golgi complex of a typical mammalian cell has 3 or 4 membranous sacs (cisternae), and those of many plant cells have about 20. The Golgi complex is differentiated into (1) a cis compartment, the receiving end, which is closest to the ER; (2) medial compartments; and (3) a trans compartment, which exports proteins to a variety of destinations. These compartments contain different enzymes and mediate distinctive functions. The N-linked carbohydrate units of glycoproteins are further modified in each of the compartments of the Golgi complex. In the cis Golgi compartment, three mannose residues are removed from the oligosaccharide chains of proteins destined for secretion or for insertion in the plasma membrane. The carbohydrate units of glycoproteins targeted to the lumen of lysosomes are further modified. In the medial Golgi compartments of some cells, two more mannose residues are removed, and two N- acetylglucosamine residues and a fucose residue are added. Finally, in the trans Golgi, another N-acetylglucosamine residue can be added, followed by galactose and sialic acid, to form a complex oligosaccharide unit. The sequence of N-linked oligosaccharide units of a glycoprotein is determined both by (1) the sequence and conformation of the protein undergoing glycosylation and by (2) the glycosyltransferases present in the Golgi compartment in which they are processed. Note that, despite all of this processing, N-glycosylated proteins have in common a pentasaccharide core. Carbohydrate processing in the Golgi complex is called terminal glycosylation to distinguish it from core glycosylation, which takes place in the ER. Tremendous structural diversification can occur as a result of the terminal glycosylation process.
PROTEIN
TARGET TO LYSOSOMES
This sorting process is perhaps best understood in the case of hydrolases destined for transport to lysosomes. Upon arrival in the Golgi complex from the ER, some as yet undetermined feature of the threedimensional structure of these hydrolases (sometimes called a "signal patch") is recognized by a phosphotransferase that catalyzes the phosphorylation of certain mannose residues in the enzymes' oligosaccharides. The presence of one or more mannose-6-phosphate residues in their N-linked oligosaccharides is the structural signal that targets these proteins to lysosomes. A receptor protein in the membrane of the Golgi complex recognizes this mannose-6-phosphate signal and binds the hydrolases so marked. Vesicles containing these receptor-hydrolase complexes bud from the trans side of the Golgi complex and make their way to sorting vesicles. Here, the receptorhydrolase complexes dissociate in a process facilitated by the lower pH within the sorting vesicles and by a phosphatase-catalyzed removal of phosphate groups from the mannose-6-phosphate residues. The receptor is returned to the Golgi complex, and vesicles containing the hydrolases bud from the sorting vesicles and move to the lysosomes. In cells treated with tunicamycin, hydrolases normally targeted for lysosomes do not reach their destination but are secreted instead, confirming that the N-linked oligosaccharide plays a key role in targeting these enzymes to lysosomes.
![]()
Formation of a
Mannose 6-Phosphate Marker. A
glycoprotein destined for delivery to lysosomes acquires a phosphate marker in
the cis Golgi compartment in a two-step process. First, a phosphotransferase
adds a phospho-N-acetylglucosamine unit to the 6-OH group of a mannose,
and then a phosphodiesterase removes the added sugar to generate a mannose
6-phosphate residue in the core oligosaccharide.
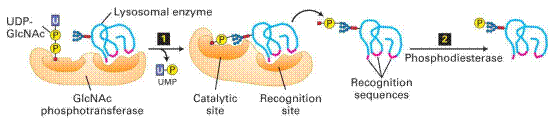
The M6P residues
that direct proteins to lysosomes are generated in the cis-Golgi by two
Golgi-resident enzymes. Step1: An Nacetylglucosamine
(GlcNAc) phosphotransferase transfers a
phosphorylated GlcNAc group to carbon atom 6 of one
or more mannose residues. Because only lysosomal enzymes contain sequences
(red) that are recognized and bound by this enzyme, phosphorylated GlcNAc groups are added specifically to lysosomal enzymes.
Step2: After release of a modified protein from the phosphotransferase, a
phosphodiesterase removes the GlcNAc group, leaving a
phosphorylated mannose residue on the lysosomal enzyme.
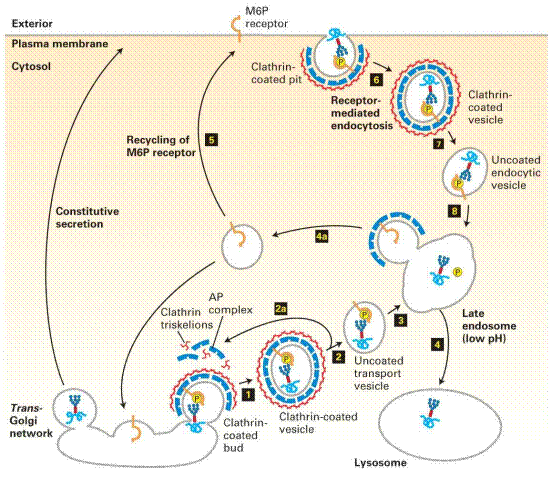
Newly synthesized
lysosomal enzymes, produced in the ER, acquire mannose 6-phosphate (M6P)
residues in the cis-Golgi. For simplicity, only one phosphorylated
oligosaccharide chain is depicted, although lysosomal enzymes typically have
many such chains. In the trans-Golgi network, proteins that bear the M6P
sorting signal interact with M6P receptors in the membrane and thereby are
directed into clathrin/AP1 vesicles (step1). The coat
surrounding released vesicles is rapidly depolymerized (step2), and the
uncoated transport vesicles fuse with late endosomes (step3). After the
phosphorylated enzymes dissociate from the M6P receptors and are
dephosphorylated, late endosomes subsequently fuse with a lysosome (step4).
Note that coat proteins and M6P receptors are recycled (steps 2a and 4a), and
some receptors are delivered to the cell surface (step5). Phosphorylated
lysosomal enzymes occasionally are sorted from the trans-Golgi to the
cell surface and secreted. These secreted enzymes can be retrieved by
receptor-mediated endocytosis (steps 68), a process that closely parallels
trafficking of lysosomal enzymes from the trans-Golgi network to
lysosomes.
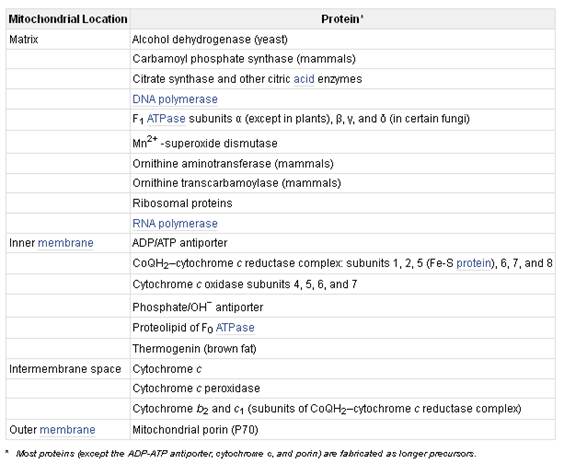
MITOCHONDRIAL TRANSPORT
Most mitochondrial proteins, however, are synthesized outside the organelle on cytosolic ribosomes that are not bound to the rough endoplasmic reticulum. The newly made proteins are released into the cytosol and are then taken up specifically into the proper organelle by binding to receptor proteins on the organelle surface that recognize specific uptake-targeting sequences in the new proteins (Table). The mitochondrion contains multiple membranes and membrane-limited spaces. Thus targeting of some proteins requires the sequential action of two targeting sequences and two membrane-bound receptor systems: one to direct the protein into the organelle, and the other to direct it into the correct organellar compartment or membrane. In general, protein uptake into mitochondria is an energy-requiring process that depends on integral proteins in the organellar membranes.
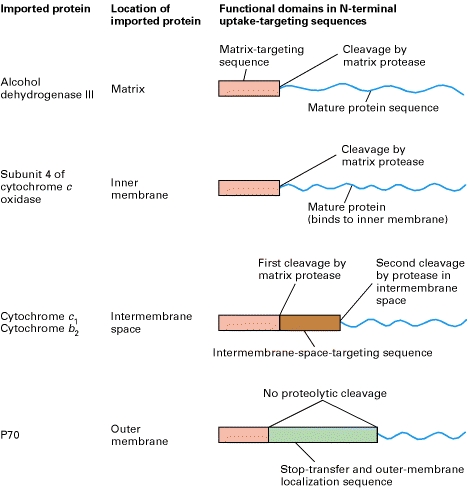
TRANSPORT TO MATRIX
As a precursor protein, with its N-terminal matrix-targeting sequence (red), emerges from cytosolic ribosomes, it binds to chaperone proteins, such as mitochondrial-import stimulating factor (MSF) and cytosolic Hsc70. These chaperones use energy released by ATP hydrolysis to keep the bound precursor in an unfolded or partially folded state.
Steps 1a, 2, and 3a: MSF binds to the matrix-targeting sequence in some precursors and uses energy released by ATP hydrolysis to keep the precursors unfolded. MSF then binds to a Tom37/Tom70 receptor on the outer membrane. The bound precursor, in turn, is delivered to a second receptor, Tom20/Tom22, which recognizes the matrix targeting sequence. Steps 1b and 3b: Other precursor proteins bind to cytosolic Hsc70, which also uses energy released by ATP hydrolysis to keep the precursors unfolded. These are delivered directly to Tom20/Tom22 receptors. Step 4: Once a precursor protein is bound to a Tom20/Tom22 receptor near a site of contact with the inner membrane, it is transported across the outer membrane through a transport channel composed of Tom40 and three smaller but essential subunits (not depicted here). Step 5: The precursor protein is then translocated across the inner membrane through another transport channel composed of several different Tim proteins. This process requires a proton-motive force (pmf), a combination of a membrane electric potential and a pH gradient, across the inner membrane. Note that translocation occurs at rare contact sites at which the inner and outer membranes appear to touch. The newly imported protein binds to the matrix chaperone Hsc70, itself bound to the Tim44 subunit of the inner-membrane transport channel. Hsc70 uses the energy of ATP hydrolysis to assist import into the matrix and to prevent aggregation or premature folding. Step 6: After Hsc70 is released, the uptake-targeting sequence is removed by a matrix protease. Step 7a: Within the matrix, some proteins fold into their mature, active conformation without the aid of a chaperone. Step 7b: Other proteins bind to the chaperonin Hsc60, which assists in the final folding in a process that requires energy derived from ATP hydrolysis.
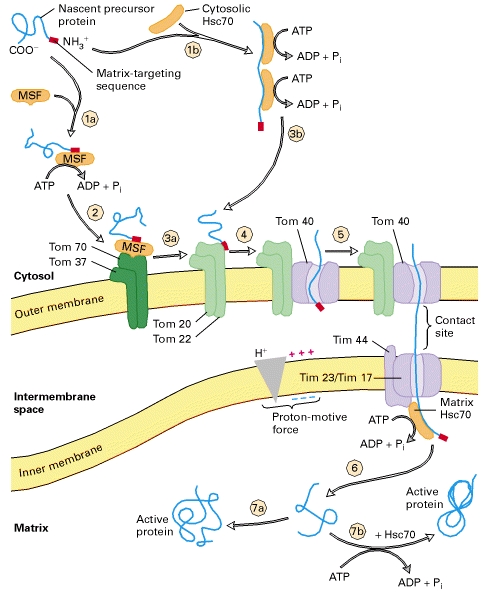
TRANSPORT TO INTERMEMBRANE SPACE
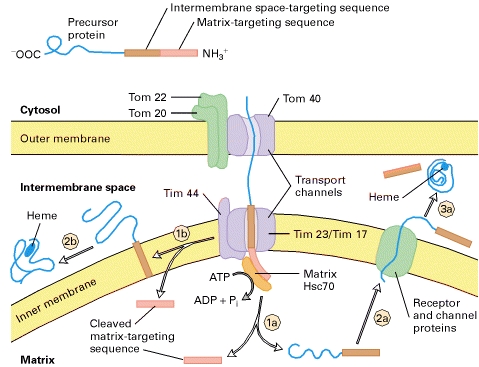
Two pathways by which different proteins are
transported from the cytosol to the mitochondrial intermembrane space. In both pathways, the precursor
protein contains two N-terminal targeting sequences (top). It is
delivered to Tom20/Tom22 receptors in the outer membrane and begins
translocation across the transport channels. Conservative pathway: The
entire precursor enters the matrix exactly as if it were a typical mitochondrial
matrix protein, and the matrix-targeting sequence (red) is cleaved by the
matrix protease (step 1a). The protein (e.g., cytochrome c1)
remains unfolded, presumably bound to matrix Hsc70 (not shown here). The
intermembrane space-targeting sequence (brown) then targets the protein to the
inner membrane by binding to a receptor (green) on the matrix side of the inner
membrane, after which the protein is translocated across the inner membrane
through an associated protein-lined transport channel into the intermembrane
space (step 2a). (Neither the receptor nor the channel proteins have been
characterized.) In the intermembrane space, the targeting sequence is cleaved
by a specific protease that is related to the ER signal peptidase, and heme is
added, enabling the cytochrome to fold into its mature configuration (step 3a). Nonconservative pathway: The matrix-targeting sequence moves
across both the outer and inner membranes, but the hydrophobic intermembrane
space-targeting sequence becomes anchored in the inner membrane. This prevents
translocation of the C-terminus of the protein (e.g., cytochrome b2)
through the inner membrane and apparently causes disassembly of the transport
channel. The targeting sequence anchored in the inner membrane diffuses away from
the translocation site (step 1b), as the rest of the protein
traverses the outer membrane into the intermembrane space, and the
matrix-targeting sequence is cleaved. Cleavage of the intermembrane
space-targeting sequence by a specific protease releases the protein to which
heme is added, followed by folding of the cytochrome into its mature
conformation (step 2b).
TRANSPORT TO MITOCHONDRIAL
MEMBRANE
Some mitochondrial proteins are targeted to the outer membrane or inner membrane, rather than to the matrix, so additional mechanisms are needed to direct these proteins to the correct submitochondrial compartment. These proteins are targeted to their destinations by a second sorting signal following the positively charged presequence that directs mitochondrial import. The targeting of proteins to the mitochondrial membranes appears to be mediated by hydrophobic stop-transfer sequences that halt translocation of the polypeptide chains through the Tim or Tom complexes, leading to their insertion into the inner or outer mitochondrial membranes, respectively
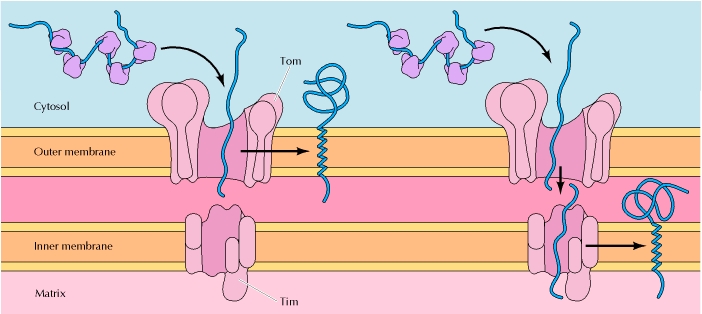
NULCEAR TRANSPORT SIGNAL SEQUENCE
Molecular communication between the nucleus and the cytosol requires the movement of macromolecules through nuclear pores. RNA molecules synthesized in the nucleus are exported to the cytosol. Ribosomal proteins synthesized on cytosolic ribosomes are imported into the nucleus and assembled into 60S and 40S ribosomal subunits in the nucleolus; completed subunits are then exported back to the cytosol. A variety of nuclear proteins (RNA and DNA polymerases, histones, topoisomerases, proteins that regulate gene expression, and so forth) are synthesized in the cytosol and imported into the nucleus. This traffic is modulated by a complex system of molecular signals and transport proteins that is gradually being elucidated.
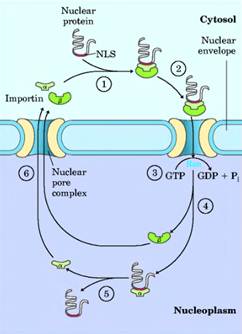
In most multicellular eukaryotes, the nuclear envelope breaks down at each cell division, and once division is completed and the nuclear envelope reestablished, the dispersed nuclear proteins must be reimported. To allow this repeated nuclear importation, the signal sequence that targets a protein to the nu- nucleusthe nuclear localization sequence, NLSis not removed after the protein arrives at its destination. An NLS, unlike other signal sequences, may be located almost anywhere along the primary sequence of the protein. NLSs can vary considerably, but many consist of four to eight amino acid residues and include several consecutive basic (Arg or Lys) residues. Nuclear importation is mediated by a number of proteins that cycle between the cytosol and the nucleus (Fig. 27-37), including importin a and j8 and a small GTPase known as Ran. A heterodimer of importin a and j8 functions as a soluble receptor for proteins targeted to the nucleus, with the a subunit binding NLS-bearing proteins in the cytosol. The complex of the NLS-bearing protein and the importin docks at a nuclear pore and is translocated through the pore by an energy- dependent mechanism that requires the Ran GTPase.
The two importin subunits separate during the translocation, and the NLS-bearing protein dissociates from importin a inside the nucleus. Importin a and j8 are then exported from the nucleus to repeat the process. How importin a remains dissociated from the many NLS-bearing proteins inside the nucleus is not yet clear.
BACTERIAL PROTEIN EXPORT
SIGNAL SEQUENCE
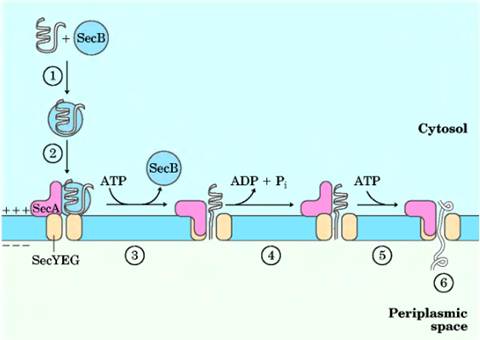
Bacteria can target proteins to their inner or outer membranes, to the periplasmic space between these membranes, or to the extracellular medium. They use signal sequences at the amino terminus of the proteins, much like those on eukaryotic proteins targeted to the ER, mitochondria, and chloroplasts. Most proteins exported from E. coli make use of the pathway shown in Figure 27-39. Following translation, a protein to be exported may fold only slowly, the amino-terminal signal sequence impeding the folding.
The soluble chaperone protein SecB binds to the protein's signal sequence or other
features of its incompletely folded structure. The bound protein is then
delivered to SecA, a protein associated with the
inner surface of the plasma membrane. SecA acts as
both a receptor and a translocating ATPase. Released from SecB
and bound to SecA, the protein is delivered to a
translocation complex in the membrane, made up of SecY,
E, and G, and is translocated stepwise through the membrane at the SecYEG complex in lengths of about 20 amino acid residues.
Each step is facilitated by the hydrolysis of ATP, catalyzed by SecA.
SECRETION PATHWAY
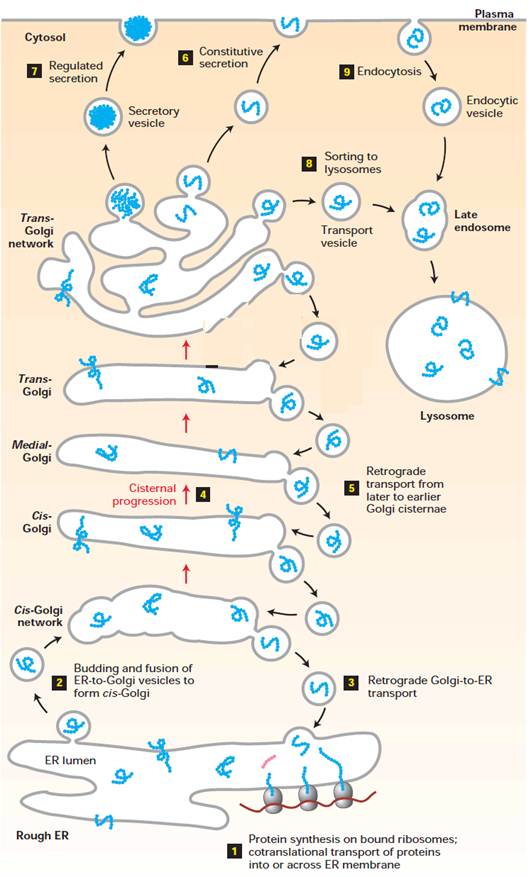
Overview of the secretory and
endocytic pathways of protein sorting. Secretory pathway: Synthesis of proteins bearing an ER signal
sequence is completed on the rough ER (1), and the newly made polypeptide chains
are inserted into the ER membrane or cross it into the lumen. Some proteins
(e.g., ER enzymes or structural proteins) remain within the ER. The remainder
are packaged into transport vesicles (2) that bud from the ER and fuse together
to form new cis-Golgi cisternae. Missorted ER-resident proteins and
vesicle membrane proteins that need to be reused are retrieved to the ER by
vesicles (3) that bud from the cis-Golgi and fuse with the ER. Each cis-Golgi
cisterna, with its protein content, physically moves from the cis to the
trans face of the Golgi complex (4) by a nonvesicular
process called cisternal progression. Retrograde transport vesicles (5) move
Golgi-resident proteins to the proper Golgi compartment. In all cells, certain
soluble proteins move to the cell surface in transport vesicles (6) and are
secreted continuously (constitutive
secretion). In certain cell types, some soluble proteins are stored in
secretory vesicles (7) and are released only after the cell receives an
appropriate neural or hormonal signal (regulated secretion). Lysosome-destined
membrane and soluble proteins, which are transported in vesicles that bud from
the trans-Golgi (8), first move to the late endosome and then to the
lysosome. Endocytic pathway: Membrane and soluble extracellular proteins
taken up in vesicles that bud from the plasma membrane (9) also can move to the
lysosome via the endosome.
COATED
VESICLES INVOLVED IN PROTEIN SORTING
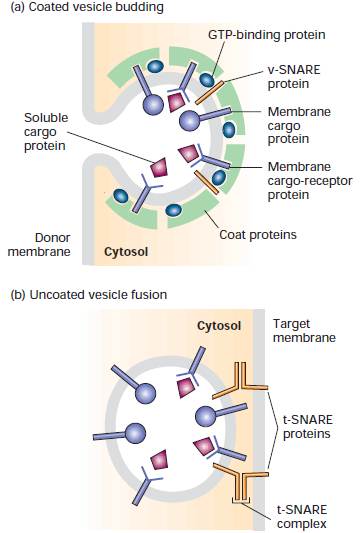
Overview of vesicle budding and fusion with a target membrane. (a) Budding is initiated by recruitment of a small GTP-binding protein to a patch of donor membrane. Complexes of coat proteins in the cytosol then bind to the cytosolic domain of membrane cargo proteins, some of which also act as receptors that bind soluble proteins in the lumen, thereby recruiting luminal cargo proteins into the budding vesicle. (b) After being released and shedding its coat, a vesicle fuses with its target membrane in a process that involves interaction of cognate SNARE proteins.
THREE TYPES OF
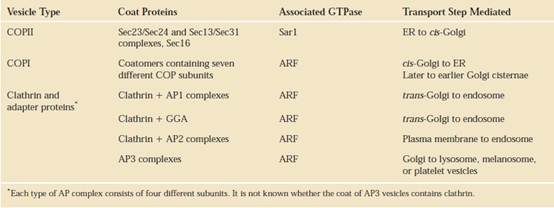
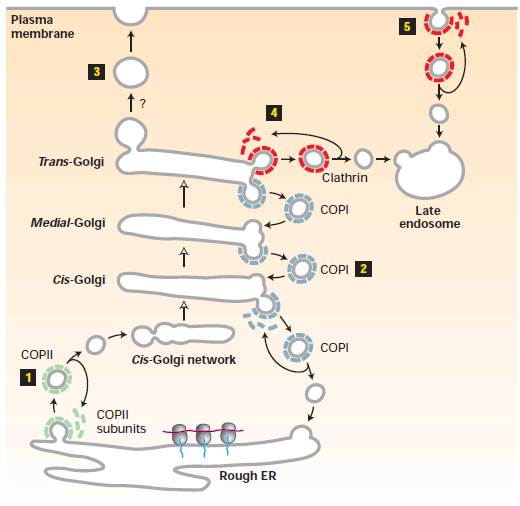
COATED VESICLES TRANSPORT PROTEINS FROM ORGANELLE TO ORGANELLE (COP stands for coat protein)
After formation of
vesicles by budding from a donor membrane, the coats depolymerize into their
subunits, which are re-used to form additional transport vesicles. COPII
vesicles (1) mediate anterograde transport from the rough ER to the cis-Golgi/cis-Golgi
network. COPI vesicles (2) mediate retrograde transport within the Golgi and
from the cis-Golgi/cis-Golgi network to the rough ER. The coat
proteins surrounding secretory vesicles (3) are not yet characterized; these
vesicles carry secreted proteins and plasma-membrane proteins from the trans-Golgi
network to the cell surface. Vesicles coated with clathrin
(red) bud from the trans-Golgi network (4) and from the plasma membrane
(5); after uncoating, these vesicles fuse with late endosomes. The coat on most
clathrin vesicles contains additional proteins not
indicated here. Note that secretory proteins move from the cis- to trans-Golgi
by cisternal progression, which is not mediated by vesicles. Question marks indicate that the nature of the coat is unknown.
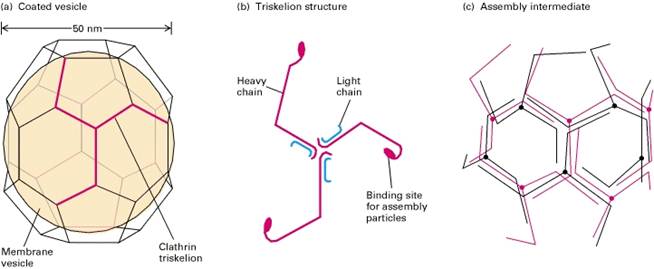
Clathrin
Typical clathrin-coated vesicles are 50 100 nm in diameter, with a membrane-bounded vesicle inside a coat composed primarily of the fibrous protein clathrin (Figure-a). Purified clathrin molecules, which have a three-limbed shape, are called triskelions from the Greek for three-legged (Figure-b). Each limb contains one clathrin heavy chain (180,000 MW) and one clathrin light chain (≈35,000 40,000 MW). There are two types of light chains, α and β, whose amino acid sequences are 60 percent identical; their functional differences are not known. Even in the absence of membrane vesicles, clathrin triskelions can polymerize to form the cage-like structure that is found around a coated vesicle. When clathrin polymerizes, it forms a polygonal lattice with an intrinsic curvature (Figure-c).
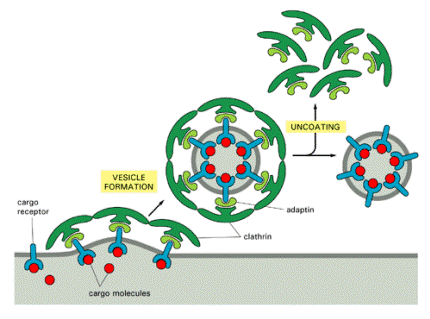
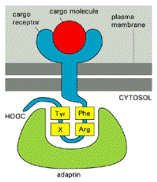
Adaptor proteins
Between the fibrous clathrin and the membrane of a clathrin-coated pit lies a 20-nm space containing assembly particles. Each particle (340,000 MW) contains one copy each of four different adapter proteins. Assembly particles bind to the globular domain at the end of each clathrin heavy chain in a triskelion) and promote the polymerization of clathrin triskelions into cages. By also binding to the cytosolic face of membrane proteins, assembly particles determine which proteins are specifically included in (or excluded from) the budding transport vesicle. Three types of assembly particles AP1, AP2, and AP3 composed of different, though related, adapter proteins, have been identified and found to mediate specific transport steps.
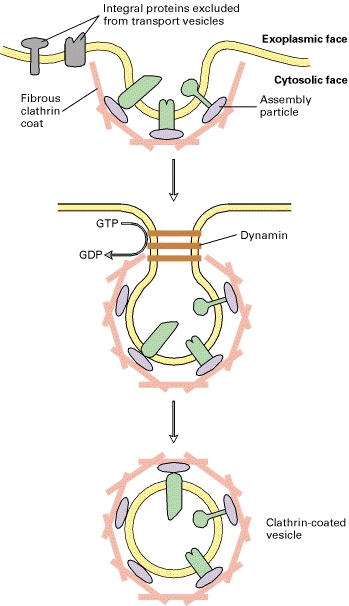
Dynamin
Essential to formation of a completed clathrin-coated pit, dynamin is an ≈900-amino-acid cytosolic protein that binds and then hydrolyzes GTP. After dynamin subunits polymerize around the neck of a pit, hydrolysis of GTP is thought to regulate contraction of the polymeric dynamin until the vesicle pinches off. Incubation of cell extracts with a derivative of GTP that cannot be hydrolyzed provides dramatic evidence for the importance of dynamin in endocytosis. Such treatment leads to accumulation of clathrin-coated pits with excessively long necks that are surrounded by polymeric dynamin but do not pinch off. Likewise, the cellular expression of mutant dynamins that cannot bind GTP blocks the formation of clathrin-coated vesicles, resulting in accumulation of similar long-necked pits encased with polymerized dynamin.
COATOMER-COATED
VESICLE
ARF is a monomeric GTPase with a fatty acid tail, and it is thought to play a crucial role in both the assembly and disassembly of coatomer coats. It is found in high concentration in the cytosol in its discharged, GDP-bound state. It seems that the donor membrane from which a coatomer-coated vesicle is to bud contains a specific guanine-nucleotide-releasing protein that causes ARF to release its GDP and bind GTP in its place (as GTP is present in much higher concentration in the cytosol than GDP). The binding of GTP is thought to cause the ARF to expose its fatty acid tail, which inserts into the lipid bilayer of the donor membrane. The tightly bound ARF now recruits coatomer subunits, which bind to it. The assembly of the coatomer coat, which consists of both the GTP-charged ARF and the coatomer proteins, pulls the membrane into a bud, which then pinches off as a coated vesicle (Figure 13-55).
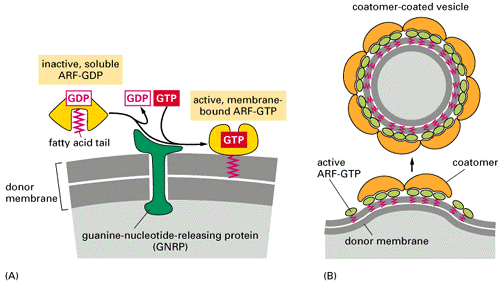
A current model of coatomer-coated vesicle formation. (A) Inactive, soluble ARF-GDP binds to a guaninenucleotide-releasing protein in the donor membrane, causing the ARF to release its GDP and bind GTP. A GTP-triggered conformational change in ARF exposes its fatty acid chain, which inserts into the donor membrane. (B) Membrane-bound, active ARF-GTP recruits coatomer subunits to the membrane. This causes the membrane to form a bud. A sub-sequent membrane-fusion event pinches off and releases the coated vesicle. The drug brefeldin A blocks coatomer-coat assembly by inhibiting the exchange reaction of GDP to GTP. This blocks coatomer-coated vesicular traffic from the ER through the Golgi apparatus, causing the Golgi apparatus to empty into the ER
When the coatomer-coated vesicle docks
with its target membrane, a specific GTPase-activating protein in the target
membrane triggers the ARF to hydrolyze its bound GTP to GDP. This is thought to
lead to a conformational change in ARF so that its fatty acid chain pops out of
the membrane, causing the vesicle's coat to disassemble and allowing membrane
fusion to proceed, as discussed later. Thus ARF can be
viewed as a protein that senses the circumstances and gives the appropriate
signal, either for coat assembly and vesicle budding or for coat disassembly
and vesicle docking, as the case may be. Most important, given a
guanine-nucleotide-releasing protein in the donor membrane and a
GTPase-activating protein in the target membrane, the direction of transport is
defined: because of its cycle of GTP hydrolysis and GDP/GTP exchange, ARF
facilitates transfer in one direction only.
Known sorting signals that direct proteins to specific transport
vesicles
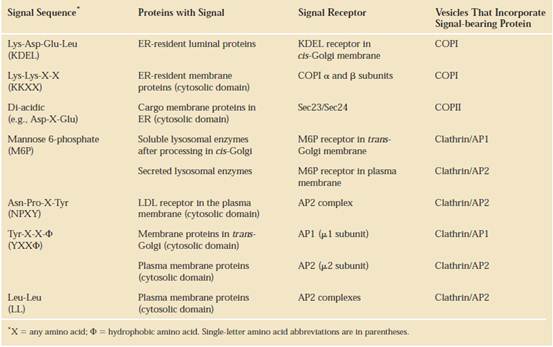
MODEL FOR DOCKING AND FUSION OF TRANSPORT VESICLES WITH THEIR TARGET
MEMBRANES.
The proteins shown in this example participate
in fusion of secretory vesicles with the plasma membrane, but similar proteins
mediate all vesicle-fusion events. Step1 : A Rab protein tethered via a lipid anchor to a secretory
vesicle binds to an effector protein complex on the plasma membrane, thereby
docking the transport vesicle on the appropriate target membrane. Step2: A
v-SNARE protein (in this case, VAMP) interacts with the cytosolic domains of
the cognate t-SNAREs (in this case, syntaxin and
SNAP-25). The very stable coiled-coil SNARE complexes that are formed hold the
vesicle close to the target membrane. Inset: Numerous noncovalent interactions
between four long a helices, two from SNAP-25 and one each from syntaxin and VAMP, stabilize the coiled-coil structure.
Step3: Fusion of the two membranes immediately follows formation of SNARE
complexes, but precisely how this occurs is not known. Step4: Following
membrane fusion, NSF in conjunction with a-SNAP protein binds to the SNARE complexes. The
NSF-catalyzed hydrolysis of ATP then drives dissociation of the SNARE
complexes, freeing the SNARE proteins for another round of vesicle fusion.
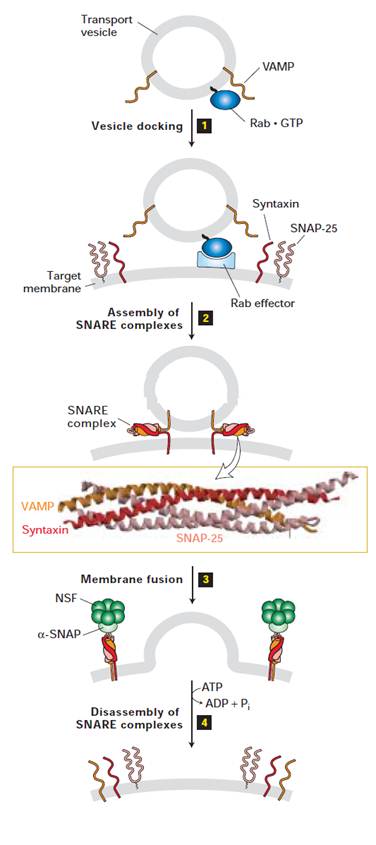
RAB PROTEINS AND VESICLE TRANSPORT
Because there are many different membrane systems in the cell, the docking process must be ighly selective. A vesicle is likely to inspect many potential target membranes before its v-SNARE finds a complementary t-SNARE. According to one view, this crucial recognition step is controlled by members of a family of monomeric GTPases called Rab proteins, which check that the fit between a v-SNARE and a t-SNARE is correct. In this view Rab proteins become attached to the surface of budding coated vesicles in the donor membrane. When a vesicle encounters the correct target membrane, the binding of v-SNARE to t-SNARE causes the vesicle to remain bound for long enough to allow the Rab protein to hydrolyze its bound GTP, which locks the vesicle onto the target membrane, readying it for subsequent fusion.
Eucaryotic cells contain many types of Rab proteins, each associated with a particular membrane-bounded organelle involved in the secretory or endocytic pathways. Each of these organelles has at least one Rab protein on its cytosolic surface. The first Rab protein (called Sec4) was discovered in yeast by selecting for mutations (called SEC mutations) that interfere with secretion. It was subsequently shown to be a component of secretory vesicles and is required for their docking at the plasma membrane; mutations that disrupt it prevent secretory vesicles from discharging their contents to the exterior. The amino acid sequences of Rab proteins are most dissimilar near their carboxyl-terminal tails, and tail-swapping experiments using genetic engineering techniques indicate that it is the tail that determines the intracellular location of each family member, presumably by enabling it to bind to a complementary guanine-nucleotide-releasing factor on the surface of a particular organelle. Before SNAREs became candidates for the organelle marker proteins that guide vesicular transport, Rab proteins were thought to play this role because of their remarkable organelle-specific distribution. It is now known, however, that at least some Rab proteins are functionally interchangeable as long as they are experimentally engineered to become localized to the new organelle. Thus they cannot be the sole explanation for the selectivity of vesicular transport.
SUBCELLULAR
LOCATIONS OF SOME RAB PROTEINS
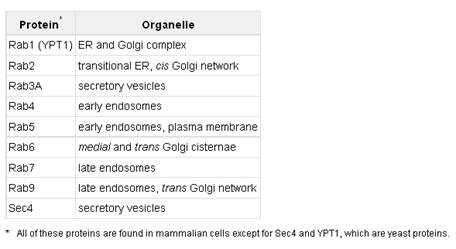
PROTEIN DEGRADATION
Protein degradation prevents the buildup of abnormal or unwanted proteins and permits the recycling of amino acids. The half-lives of eukaryotic proteins vary from 30 seconds to many days. Most proteins turn over rapidly relative to the lifetime of a cell, although a few (such as hemoglobin) can last for the life of the cell (about 110 days for an erythrocyte). Rapidly degraded proteins include those that are defective because of incorrectly inserted amino acids or because of damage accumulated during normal functioning. And enzymes that act at key regulatory points in metabolic pathways often turn over rapidly.
Defective proteins and those with characteristically short half-lives are generally degraded in both bacterial and eukaryotic cells by selective ATP-dependent cytosolic systems. A second system in vertebrates, operating in lysosomes, recycles the amino acids of membrane proteins, extracellular proteins, and proteins with characteristically long half-lives.
In E. coli, many proteins are degraded by an ATP-dependent protease called Lon (the name refers to the "long form" of proteins, observed only when this protease is absent). The protease is activated in the presence of defective proteins or those slated for rapid turnover; two ATP molecules are hydrolyzed for every peptide bond cleaved. The precise role of this ATP hydrolysis is not yet clear. Once a protein has been reduced to small inactive peptides, other ATP-independent proteases complete the degradation process.
The ATP-dependent pathway in eukaryotic cells is quite different, involving the protein ubiquitin, which, as its name suggests, occurs throughout the eukaryotic kingdoms. One of the most highly conserved proteins known, ubiquitin G6 amino acid residues) is essentially identical in organisms as different as yeasts and humans. Ubiquitin is covalently linked to proteins slated for destruction via an ATP-dependent pathway involving three separate enzymes (El, E2, and E3).
Ubiquitinated proteins are degraded by a large complex known as the 26S proteasome (Afr 2.5 X 106) (Fig. 27-42). The proteasome consists of two copies each of at least 32 different subunits, most of which are highly conserved from yeasts to humans. The proteasome contains two main types of subcomplexes, a barrel-like core particle and regulatory particles on either end of the barrel. The 2OS core particle consists of four rings; the outer rings are formed from seven a subunits, and the inner rings from seven j8 subunits. Three of the seven subunits in each j8 ring have protease activities, each with different substrate specificities. The stacked rings of the core particle form the barrel-like structure within which target proteins are degraded. The 19S regulatory particle on each end of the core particle contains 18 subunits, including some that recognize and bind to ubiquitinated proteins. Six of the subunits are ATPases that probably function in unfolding the ubiquitinated proteins and translocating the unfolded polypeptide into the core particle for degradation. Although we do not yet understand all the signals that trigger ubiquitination, one simple signal has been found. For many proteins, the identity of the first residue that remains after removal of the amino-terminal Met residue, and any other posttranslational proteolytic processing of the amino-terminal end, has a profound influence on half-life. These amino-terminal signals have been conserved over billions of years of evolution, and are the same in bacterial protein degradation systems and in the human ubiquitination pathway.
Ubiquitin-dependent proteolysis is as important for the regulation of cellular processes as for the elimination of defective proteins. Many proteins required at only one stage of the eukaryotic cell cycle are rapidly degraded by the ubiquitin-dependent pathway after completing their function. The same pathway also processes and presents class I MHC antigens. Ubiquitin-dependent destruction of cyclin is critical to cell-cycle regulation. The E2 and E3 components of the ubiquitination cascade pathway are in fact two large families of proteins. Different E2 and E3 enzymes exhibit different specificities for target proteins and thus regulate different cellular processes. Some E2 and E3 enzymes are highly localized in certain cellular compartments, reflecting a specialized function.
Three-step cascade pathway by which ubiquitin is attached to a protein.Two different enzyme-ubiquitin intermediates are involved. The free carboxyl group of ubiquitin's carboxyl-terminal Gly residue is ultimately linked through an amide (isopeptide) bond to an e-amino group of a Lys residue of the target protein. Additional cycles produce polyubiquitin, a covalent polymer of ubiquitin subunits that targets the attached protein for destruction in eukaryotes.
Linkage and Selective Proteolysis
Ensure All-or-None Assembly
Many proteins are present in large complexes with other proteins. This requires that the protein bind to several other proteins at the same time. It is crucial for the cell that each protein complex form efficiently and that the formation of partial complexes, which can interfere with the function of complete complexes, be kept to a minimum. There must be mechanisms, therefore, for ensuring that assembly is an all-or-none process.
One important mechanism relies on the phenomenon of linkage, which we described earlier. Because of linkage, if a ligand changes the shape of an allo-steric protein so that the protein binds a second ligand more tightly, the second ligand must similarly increase the affinity of the protein for the first ligand. The same principle applies to protein-protein interactions. When two proteins bind to each other, they often increase the affinity of one of the partners for a third protein. Because of linkage, the complex of all three proteins will be much more stable than a complex containing only two. A mechanism of this type can produce all-or-none assembly. Even if an all-or-none assembly mechanism drives the formation of complete protein complexes, unless the cell contains exactly the right proportions of each protein in the complex, unassembled proteins will be left over. In fact, cells do not always produce their components in precise amounts and are instead able to degrade selectively any protein component that is left unassembled. Cells therefore require a sophisticated system to identify abnormally assembled proteins and destroy them. Indeed, the eucaryotic cell contains an elaborate set of proteins that enables such incomplete assemblies to be selectively directed to its protein-degradation machinery, as we now discuss.
Linkage
facilitates an efficient all-or-none assembly of protein complexes. As indicated, proteins X and Y each induce an
allosteric shape change in a third protein (shown in blue) that helps
the other protein to bind. As a result, the complex of all three proteins may
be the only one that is strong enough to exist in the cell, resulting
effectively in all-or-none assembly.
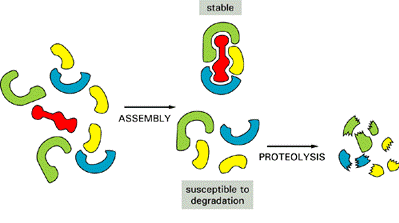
Proteolysis of the extra components of a protein complex prevents them from accumulating in a cell. The degradation shown here requires that an unassembled protein be recognized by enzymes that covalently add ubiquitin to it, as discussed in the text.
UBIQUITIN
PATHWAY
One function of intracellular proteolytic mechanisms is to recognize and eliminate unassembled proteins, as just described. Another is to dispose of damaged or misfolded proteins. Yet another is to confer short half-lives on certain normal proteins whose concentrations must change promptly withalterations in the state of a cell; many of these short-lived proteins are degraded rapidly at all times, while others, most notably the cyclins, are stable until they are suddenly degraded at one particular point in the cell cycle. Although here we mainly discuss how proteins are degraded in the cytosol, important degradation pathways also operate in the endoplasmic reticulum (ER) and, in lysosomes. Most of the proteins that are degraded in the cytosol are delivered to large protein complexes called proteasomes, which are present in many copies and are dispersed throughout the cell. Each proteasome consists of a central cylinder formed from multiple distinct proteases, whose active sites are thought to face an inner chamber. Each end of the cylinder is "stoppered" by a large protein complex formed from at least 10 types of polypeptides, some of which hydrolyze ATP. These protein stoppers are thought to select the proteins for destruction by binding to them and feeding them into the inner chamber of the cylinder, where multiple proteases degrade the proteins to short peptides that are then released.
Proteasomes act on proteins that have been specifically marked for destruction by the covalent attachment of a small protein called ubiquitin. Ubiquitin exists in cells either free or covalently linked to proteins. Most ubiquinated proteins have been tagged for degradation. (Some long-lived proteins such as histones are also ubiquinated, but in these cases the function of ubiquitin is not understood.) Different ubiquitin-dependent proteolytic pathways employ structurally similar but distinct ubiquitin-conjugating enzymes that are associated with recognition subunits that direct them to proteins carrying a particular degradation signal. The conjugating enzyme adds ubiquitin to a lysine residue of a target protein and thereafter adds a series of additional ubiquitin moieties, forming a multiubiquitin chain that is thought to be recognized by a specific receptor protein in the proteasome. Denatured or misfolded proteins, as well as proteins containing oxidized or otherwise abnormal amino acids, are recognized and degraded by ubiquitin-dependent proteolytic systems. The ubiquitin-conjugating enzymes presumably recognize signals that are exposed on these proteins as a result of their misfolding or chemical damage; such signals are likely to include amino acid sequences or conformational motifs that are buried and therefore inaccessible in the normal counterparts of these proteins. A proteolytic pathway that recognizes and destroys abnormal proteins must be able to distinguish between completed proteins that have "wrong" conformations and the many growing polypeptides on ribosomes (as well as polypeptides just released from ribosomes) that have not yet achieved their normal folded conformation.
That this is not a trivial problem can be demonstrated experimentally: if puromycin - an inhibitor of protein synthesis - is added to cells, the prematurely terminated proteins that are formed are rapidly degraded by a ubiquitin-dependent pathway. One possibility is that the normally forming proteins are temporarily protected by the translation machinery or by chaperone molecules. Another is that nascent and newly completed proteins are actually vulnerable to proteolysis but manage to fold up into their native conformations fast enough to escape being targeted for destruction by proteolysis.
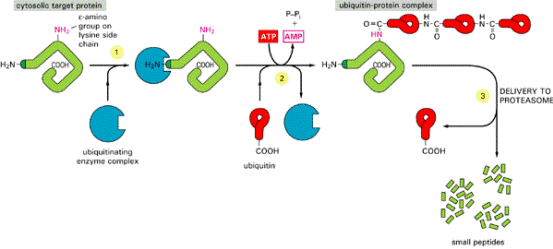
Ubiquitin-dependent
protein degradation. In step 1
a target protein (containing a degradation signal) is recognized by the
ubiquitinating enzyme complex. Then, in step 2 a repeated series of biochemical
reactions joins ubiquitin molecules together to produce a multiubiquitin chain
attached to the epsilon-amino group of a lysine side chain in the target
protein. Finally, in step 3 the proteasome cuts the target protein into a
series of small fragments.
N-END RULE AND THE PEST HYPOTHESIS
A final control
affecting the amount of gene product in a cell is control of the rate at which
proteins degrade. The normal life spans of proteins vary greatly. For example,
some proteins last longer than a cell cycle, whereas others may be broken down
in minutes. Several models have been suggested for control of protein
degradation, including the N-end rule
and the PEST hypothesis.
According to the N-end rule, the amino acid at the amino-, or N-terminal, end of a protein is a signal to proteases that control the average length of life of a protein. In recent experiments, the life span of the b-galactosidase protein was determined with almost complete predictability based on its modified N-terminal amino acid. Protein life spans range from two minutes for those with N-terminal arginine to greater than twenty hours for those with Nterminal methionine or five other amino acids.
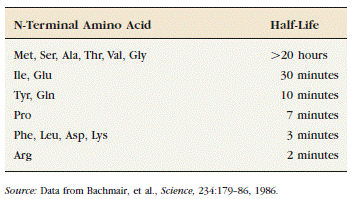
According to the PEST hypothesis, protein degradation is determined by regions rich in one of four amino acids: proline, glutamic acid, serine, and threonine. (The one-letter abbreviations of these four amino acids are P, E, S, and T, respectively.) Proteins that have these regions tend to degrade in less than 2 hours. In one study of thirty-five proteins with half-lives of between 20 and 220 hours, only three contained a PEST region. We see that not only are different proteins programmed to survive for varying lengths of time in the cell, but that programming seems to be based on the N-terminal amino acid as well as various regions rich in the PEST amino acids.
LYSOSOMAL PROTEOLYSIS
The other major pathway of protein degradation in eukaryotic cells involves the uptake of proteins by lysosomes. Lysosomes are membrane-enclosed organelles that contain an array of digestive enzymes, including several proteases (see Chapter 9). They have several roles in cell metabolism, including the digestion of extracellular proteins taken up by endocytosis as well as the gradual turnover of cytoplasmic organelles and cytosolic proteins.
The containment of proteases and other digestive enzymes within lysosomes prevents uncontrolled degradation of the contents of the cell. Therefore, in order to be degraded by lysosomal proteolysis, cellular proteins must first be taken up by lysosomes. One pathway for this uptake of cellular proteins, autophagy, involves the formation of vesicles (autophagosomes) in which small areas of cytoplasm or cytoplasmic organelles are enclosed in membranes derived from the endoplasmic reticulum (Figure 7.41). These vesicles then fuse with lysosomes, and the degradative lysosomal enzymes digest their contents. The uptake of proteins into autophagosomes appears to be nonselective, so it results in the eventual slow degradation of long-lived cytoplasmic proteins.
However, not all protein uptake by lysosomes is nonselective. For
example, lysosomes are able to take up and degrade certain cytosolic proteins
in a selective manner as a response to cellular starvation. The proteins
degraded by lysosomal proteases under these conditions contain amino acid
sequences similar to the broad consensus sequence Lys-Phe-Glu-Arg-Gln, which presumably targets
them to lysosomes. A member of the Hsp70 family of molecular chaperones is also
required for the lysosomal degradation of
these proteins, presumably acting to unfold the polypeptide chains during their
transport across the lysosomal membrane. The proteins susceptible to degradation by this pathway are thought to be
normally long-lived but dispensable proteins. Under starvation conditions,
these proteins are sacrificed to provide amino acids and energy, allowing some
basic metabolic processes to continue.
************